1NK4
 
 | |
4ZH1
 
 | | Complement factor H in complex with the GM1 glycan | | Descriptor: | Complement C3, Complement factor H, GLYCEROL, ... | | Authors: | Blaum, B.S, Stehle, T. | | Deposit date: | 2015-04-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Complement Factor H and Simian Virus 40 bind the GM1 ganglioside in distinct conformations.
Glycobiology, 26, 2016
|
|
5QAY
 
 | | OXA-48 IN COMPLEX WITH COMPOUND 32 | | Descriptor: | 1,2-ETHANEDIOL, 3-(1-methylpyrrol-2-yl)benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
4F78
 
 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Minasov, G, Egorova, O, Di Leo, R, Kudritska, M, Yim, V, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5QBE
 
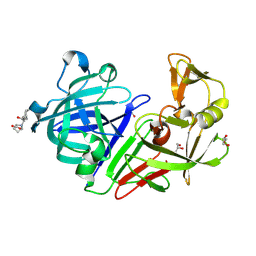 | | Crystal structure of Endothiapepsin-FRG175 complex | | Descriptor: | 1-[(1R)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]-L-proline, ACETATE ION, Endothiapepsin, ... | | Authors: | Huschmann, F. | | Deposit date: | 2017-08-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of Endothiapepsin
To be published
|
|
3BE0
 
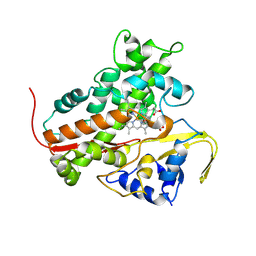 | | The Role of Asn 242 in P450cin | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, P450cin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Meharenna, Y.T, Poulos, T.L. | | Deposit date: | 2007-11-15 | | Release date: | 2008-02-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The critical role of substrate-protein hydrogen bonding in the control of regioselective hydroxylation in p450cin
J.Biol.Chem., 283, 2008
|
|
5QRE
 
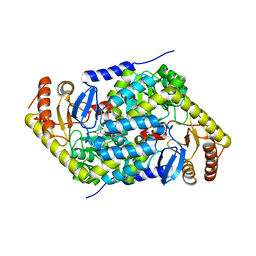 | | PanDDA analysis group deposition -- Crystal Structure of human ALAS2A in complex with Z117233350 | | Descriptor: | 3-ethyl-5-methyl-N-(5-methyl-1,2-oxazol-3-yl)-1,2-oxazole-4-carboxamide, 5-aminolevulinate synthase, erythroid-specific, ... | | Authors: | Bezerra, G.A, Foster, W, Bailey, H, Shrestha, L, Krojer, T, Talon, R, Brandao-Neto, J, Douangamath, A, Nicola, B.B, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Brennan, P.E, Yue, W.W. | | Deposit date: | 2019-05-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6IJL
 
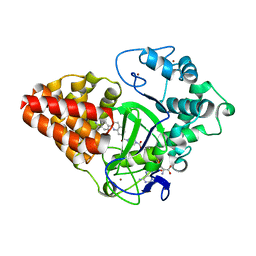 | |
5CZ7
 
 | |
5D0X
 
 | | Yeast 20S proteasome beta5-T1S mutant in complex with Bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
7FBS
 
 | | structure of a channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-[2-[(2R)-2-oxidanyl-3-(propylamino)propoxy]phenyl]-3-phenyl-propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D.J, Catterall, W.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-09-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Open-state structure and pore gating mechanism of the cardiac sodium channel.
Cell, 184, 2021
|
|
3B6Z
 
 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | Descriptor: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2UZL
 
 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 4-{5-[(Z)-(2-IMINO-4-OXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]FURAN-2-YL}-2-(TRIFLUOROMETHYL)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3BJA
 
 | |
9C3H
 
 | | Structure of the CNOT3-bound human 80S ribosome with tRNA-ARG in the P-site. | | Descriptor: | 18S rRNA, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 28S rRNA, ... | | Authors: | Erzberger, J.P, Cruz, V.E. | | Deposit date: | 2024-06-01 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Specific tRNAs promote mRNA decay by recruiting the CCR4-NOT complex to translating ribosomes.
Science, 386, 2024
|
|
1NKB
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER THREE ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP, DGTP, AND DTTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
2VJ2
 
 | | Human Jagged-1, domains DSL and EGFs1-3 | | Descriptor: | D-MALATE, JAGGED-1 | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Handford, P.A, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4H2J
 
 | |
4FGY
 
 | | Identification of a unique PPAR ligand with an unexpected binding mode and antibetic activity | | Descriptor: | (4R,6S,8S,12R,14R,16Z,18R,19R,20S,21S)-19,21-dihydroxy-22-{(2S,2'R,5S,5'S)-5'-[(1R)-1-hydroxyethyl]-2,5'-dimethyloctahydro-2,2'-bifuran-5-yl}-4,6,8,12,14,18,20-heptamethyl-9,11-dioxodocos-16-enoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, R, Li, Y. | | Deposit date: | 2012-06-05 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Identification of the antibiotic ionomycin as an unexpected peroxisome proliferator-activated receptor Gamma (PPAR-gamma) ligand with a unique binding mode and effective glucose-lowering activity in a mouse model of diabetes.
Diabetologia, 56, 2013
|
|
4NO1
 
 | | yCP in complex with Z-Leu-Leu-Leu-B(OH)2 | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(1R)-1-(dihydroxyboranyl)-3-methylbutyl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NS0
 
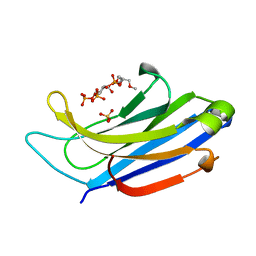 | | The C2A domain of Rabphilin 3A in complex with PI(4,5)P2 | | Descriptor: | Rabphilin-3A, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Guillen, J, Ferrer-Orta, C, Buxaderas, M, Perez-sanchez, D, Guerrero-Valero, M, Luengo-Gil, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Verdaguer, N, Corbalan-Garcia, S. | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3N2P
 
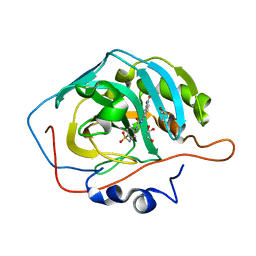 | | Crystal structure of human carbonic anhydrase II in complex with a benzenesulfonamide inhibitor | | Descriptor: | 4-{[(3-nitrophenyl)carbamoyl]amino}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, McKenna, R. | | Deposit date: | 2010-05-18 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency.
Chem.Commun.(Camb.), 46, 2010
|
|
4A6L
 
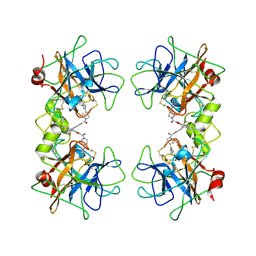 | | beta-tryptase inhibitor | | Descriptor: | 1-{3-[1-({5-[(2-fluorophenyl)ethynyl]furan-2-yl}carbonyl)piperidin-4-yl]phenyl}methanamine, TRYPTASE ALPHA/BETA-1 | | Authors: | Mathieu, M, Maignan, S. | | Deposit date: | 2011-11-04 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Library Design and the Discovery of a Potent and Selective Mast Cell Beta-Tryptase Inhibitor as an Oral Therapeutic Agent.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6I01
 
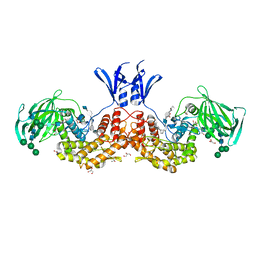 | | Structure of human D-glucuronyl C5 epimerase in complex with substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4NO6
 
 | | yCP in complex with Z-Leu-Leu-Leu-vinylsulfone | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(3S)-5-methyl-1-(methylsulfonyl)hexan-3-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
