7BEI
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-150 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEJ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
3F5W
 
 | | KcsA Potassium channel in the open-inactivated state with 32 A opening at T112 | | Descriptor: | Antibody heavy chain, Antibody light chain, POTASSIUM ION, ... | | Authors: | Cuello, L.G, Jogini, V, Cortes, D.M, Perozo, E. | | Deposit date: | 2008-11-04 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | KcsA Potassium channel in the open-inactivated state with 32 A opening at T112
TO BE PUBLISHED
|
|
6M2R
 
 | | X-ray structure of a functional Drosophila dopamine transporter in L-norepinephrine bound form | | Descriptor: | Antibody fragment 9D5 Light chain, Antibody fragment 9D5 heavy chain, CHLORIDE ION, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6M0Z
 
 | | X-ray structure of Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in L-norepinephrine bound form | | Descriptor: | Antibody fragment (Fab) 9D5 Light chain, Antibody fragment (Fab) 9D5 heavy chain, CHLORIDE ION, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
8HPT
 
 | | Structure of C5a-pep bound mouse C5aR1 in complex with Go | | Descriptor: | Antibody fragment ScFv16, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saha, S, Maharana, J, Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
3HPL
 
 | | KcsA E71H-F103A mutant in the closed state | | Descriptor: | Antibody Fab heavy chain, Antibody fab light chain, POTASSIUM ION, ... | | Authors: | Cuello, L.G, Jogini, V, Cortes, D.M, Perozo, E. | | Deposit date: | 2009-06-04 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the coupling between activation and inactivation gates in K(+) channels.
Nature, 466, 2010
|
|
8J7E
 
 | | Crystal structure of BRIL in complex with 1b3 Fab | | Descriptor: | Antibody 1b3 Fab Heavy chain, Antibody 1b3 Fab Light chain, Soluble cytochrome b562 | | Authors: | Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
3IGA
 
 | | Potassium Channel KcsA-Fab complex in Li+ and K+ | | Descriptor: | Antibody Fab Fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Thompson, A.N, Ilsoo, K, Panosian, T.D, Iverson, T.M, Allen, T.W, Nimigean, C.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4XMN
 
 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XMM
 
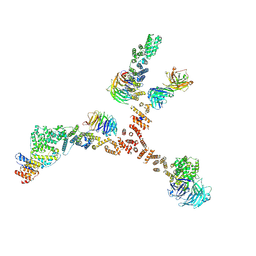 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
5GUX
 
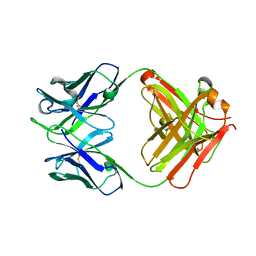 | | Cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with xenon | | Descriptor: | Antibody fab fragment heavy chain, Antibody fab fragment light chain, CALCIUM ION, ... | | Authors: | Ishii, S, Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8SCX
 
 | |
6PA0
 
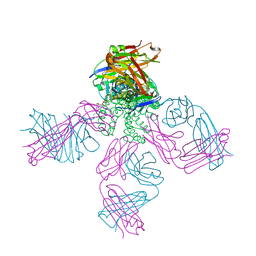 | | Structure of the G77A mutant in Sodium Chloride | | Descriptor: | Antibody HEAVY fragment, Antibody LIGHT fragment, DIACYL GLYCEROL, ... | | Authors: | Cuello, L.G, Guan, L. | | Deposit date: | 2019-06-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1QKZ
 
 | |
8JZZ
 
 | | Structure of human C5a-desArg bound human C5aR1 in complex with Go | | Descriptor: | Antibody fragment ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-07-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8E1M
 
 | |
7WGD
 
 | |
6BY2
 
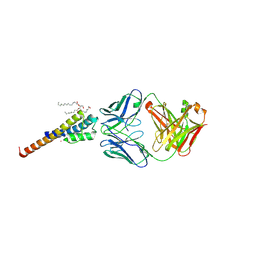 | | Closed and deep-inactivated conformation of KcsA-T75A mutant | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain, DIACYL GLYCEROL, ... | | Authors: | Labro, A.J, Cortes, D.M, Tilegenova, C, Cuello, L.G. | | Deposit date: | 2017-12-19 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inverted allosteric coupling between activation and inactivation gates in K+channels.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BY3
 
 | | Open and conductive conformation of KcsA-T75A mutant | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain, DIACYL GLYCEROL, ... | | Authors: | Labro, A.J, Cortes, D.M, Tilegenova, C, Cuello, L.G. | | Deposit date: | 2017-12-19 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Inverted allosteric coupling between activation and inactivation gates in K+channels.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4B53
 
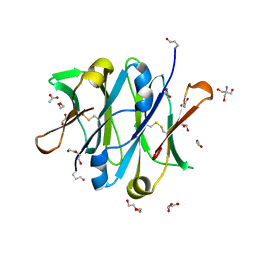 | | Crystal structure of the isolated IgG4 CH3 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Davies, A.M, Rispens, T, den Bleker, T.H, McDonnell, J.M, Gould, H.J, Aalberse, R.C, Sutton, B.J. | | Deposit date: | 2012-08-02 | | Release date: | 2012-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human Igg4 C(H)3 Dimer Reveals the Role of Arg409 in the Mechanism of Fab-Arm Exchange.
Mol.Immunol., 54, 2012
|
|
7P00
 
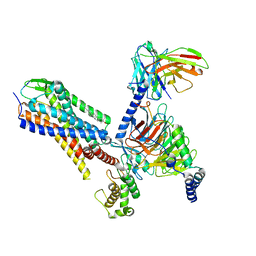 | | Human Neurokinin 1 receptor (NK1R) substance P Gq chimera (mGsqi) complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Thom, C, Ehrenmann, J, Vacca, S, Waltenspuhl, Y, Schoppe, J, Medalia, O, Pluckthun, A. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structures of neurokinin 1 receptor in complex with G q and G s proteins reveal substance P binding mode and unique activation features.
Sci Adv, 7, 2021
|
|
7P02
 
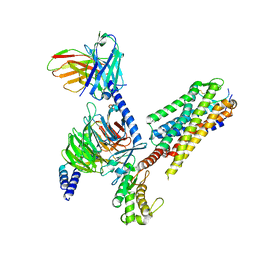 | | Human Neurokinin 1 receptor (NK1R) substance P Gs complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Thom, C, Ehrenmann, J, Vacca, S, Waltenspuhl, Y, Schoppe, J, Medalia, O, Pluckthun, A. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structures of neurokinin 1 receptor in complex with G q and G s proteins reveal substance P binding mode and unique activation features.
Sci Adv, 7, 2021
|
|
6F76
 
 | | Antibody derived (Abd-8) small molecule binding to KRAS. | | Descriptor: | 4-(2,3-dihydro-1,4-benzodioxin-5-yl)-~{N}-[3-[(dimethylamino)methyl]phenyl]-2-methoxy-aniline, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Bery, N, Cruz-Migoni, A, Quevedo, C.E, Phillips, S.V.E, Carr, S, Rabbitts, T.H. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions.
Elife, 7, 2018
|
|
7MNO
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) I656V mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.73 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
