7FD3
 
 | | IL-1RAPL2 TIR domain | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
5SRJ
 
 | |
5SOQ
 
 | |
4PK2
 
 | | tubulin acetyltransferase complex with bisubstrate analog | | Descriptor: | ACETYL-SER-(N-PROPANOYL-LYS)-ASP--THR-NH2 PEPTIDE, Alpha-tubulin N-acetyltransferase 1, COENZYME A | | Authors: | Szyk, A, Roll-Mecak, A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase.
Cell, 157, 2014
|
|
4AF3
 
 | | Human Aurora B Kinase in complex with INCENP and VX-680 | | Descriptor: | AURORA KINASE B, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, INNER CENTROMERE PROTEIN | | Authors: | Elkins, J.M, Vollmar, M, Wang, J, Picaud, S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S. | | Deposit date: | 2012-01-16 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Human Aurora B in Complex with Incenp and Vx-680.
J.Med.Chem., 55, 2012
|
|
5SOI
 
 | |
5SPA
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894417 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPB
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894404 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3D44
 
 | | Crystal structure of HePTP in complex with a dually phosphorylated Erk2 peptide mimetic | | Descriptor: | CHLORIDE ION, GLYCEROL, Mitogen-activated protein kinase 1 peptide, ... | | Authors: | Critton, D.A, Tortajada, A, Page, R. | | Deposit date: | 2008-05-13 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of substrate recognition by hematopoietic tyrosine phosphatase.
Biochemistry, 47, 2008
|
|
1SF6
 
 | | BINDING OF N,N',N"-TRIACETYLCHITOTRIOSE TO HEW LYSOZYME: A POWDER DIFFRACTION STUDY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Von Dreele, R.B. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-13 | | Method: | POWDER DIFFRACTION | | Cite: | Binding of N-acetylglucosamine oligosaccharides to hen egg-white lysozyme: a powder diffraction study.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3QZG
 
 | | CDK2 in complex with inhibitor JWS-6-76 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4,6-diamino-1,3,5-triazin-2-yl)-4-fluorophenol, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-06 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3D4Y
 
 | | GOLGI MANNOSIDASE II complex with mannoimidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
5SFE
 
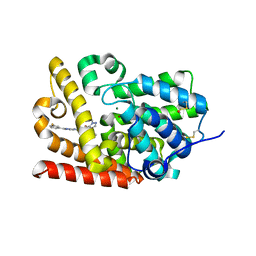 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(nc1nc(cn1cc2)c3cc(ccc3)F)NC(c4c(cnn4C)C(N(C)C)=O)=O, micromolar IC50=0.011191 | | Descriptor: | MAGNESIUM ION, N~5~-[(2P,4S)-2-(3-fluorophenyl)imidazo[1,2-a]pyrimidin-7-yl]-N~4~,N~4~,1-trimethyl-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Gobbi, L, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3QTW
 
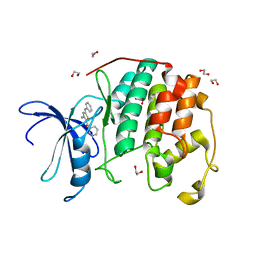 | | CDK2 in complex with inhibitor RC-2-13 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, [4-amino-2-(phenylamino)-1,3-thiazol-5-yl](pyridin-3-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3OYE
 
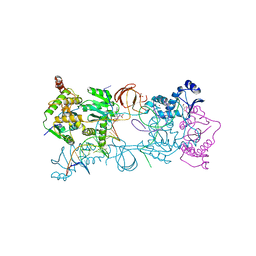 | |
3CQB
 
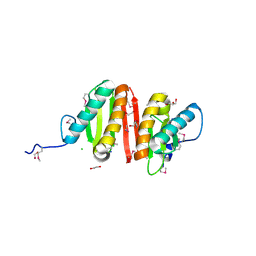 | | Crystal structure of heat shock protein HtpX domain from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-02 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of heat shock protein HtpX domain from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
4PPS
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with an A-CD ring estrogen derivative | | Descriptor: | (1S,3aR,5R,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
5RHP
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z2856434938 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GO3
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1198162455 | | Descriptor: | 1-(1-ethyl-1H-pyrazol-3-yl)-N-methylmethanamine, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
4KPE
 
 | | Novel fluoroquinolones in complex with topoisomerase IV from S. pneumoniae and E-site G-gate | | Descriptor: | (7aR,8R)-8-amino-4-cyclopropyl-12-fluoro-1-oxo-4,7,7a,8,9,10-hexahydro-1H-pyrrolo[1',2':1,7]azepino[2,3-h]quinoline-2-carboxylic acid, DNA topoisomerase 4 subunit A, DNA topoisomerase 4 subunit B, ... | | Authors: | Laponogov, I, Pan, X.-S, Vesekov, D.A, Cirz, R.T, Wagman, A.S, Moser, H.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
7GO6
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1203586731 | | Descriptor: | 1-[(4S)-imidazo[1,2-a]pyridin-7-yl]methanamine, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
3QQH
 
 | | CDK2 in complex with inhibitor L2-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-amino-6-(phenylamino)-1,3,5-triazin-2-yl]-4-chlorophenol, Cyclin-dependent kinase 2, ... | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
2Q89
 
 | | Crystal structure of EhuB in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
6ELE
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, fAB heavy chain, ... | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
3CWZ
 
 | | Structure of RAB6(GTP)-R6IP1 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab6-interacting protein 1, ... | | Authors: | Recacha, R, Houdusse, A, Goud, B, Khan, A.R. | | Deposit date: | 2008-04-23 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain.
Structure, 17, 2009
|
|
