3HVY
 
 | |
2A3U
 
 | | Crystal structure of sulbactam bound to E166A variant of SHV-1 beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, ... | | Authors: | Padayatti, P.S, Helfand, M.S, Totir, M.A, Carey, M.P, Carey, P.R, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2005-06-27 | | Release date: | 2005-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | High Resolution Crystal Structures of the trans-Enamine Intermediates Formed by Sulbactam and Clavulanic Acid and E166A SHV-1 {beta}-Lactamase.
J.Biol.Chem., 280, 2005
|
|
1DCI
 
 | | DIENOYL-COA ISOMERASE | | Descriptor: | 1,2-ETHANEDIOL, DIENOYL-COA ISOMERASE, MAGNESIUM ION, ... | | Authors: | Modis, Y, Filppula, S.A, Novikov, D, Norledge, B, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 1998-02-13 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of dienoyl-CoA isomerase at 1.5 A resolution reveals the importance of aspartate and glutamate sidechains for catalysis.
Structure, 6, 1998
|
|
1VPH
 
 | |
1VME
 
 | |
2JIW
 
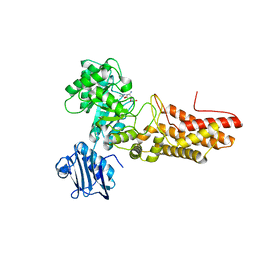 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine | | Descriptor: | N-[(1S,2R,5R,6R)-2-AMINO-5,6-DIHYDROXY-4-(HYDROXYMETHYL)CYCLOHEX-3-EN-1-YL]ACETAMIDE, O-GLCNACASE BT_4395 | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A 1-Acetamido Derivative of 6-Epi-Valienamine: An Inhibitor of a Diverse Group of Beta-N-Acetylglucosaminidases.
Org.Biomol.Chem., 5, 2007
|
|
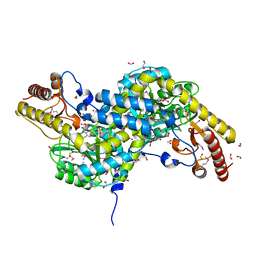
1VP4
 
 | |
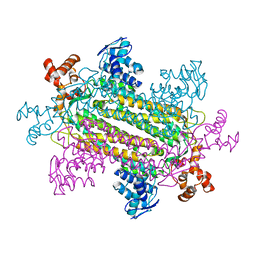
1J3U
 
 | |
5I8H
 
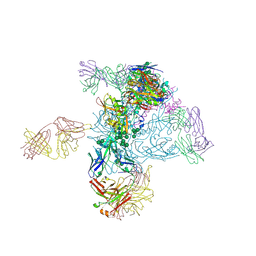 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer in Complex with V3 Loop-targeting Antibody PGT122 Fab and Fusion Peptide-targeting Antibody VRC34.01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP.664 gp120, ... | | Authors: | Xu, K, Zhou, T, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.301 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
1EJB
 
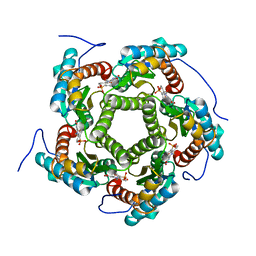 | | LUMAZINE SYNTHASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4-DIHYDROXYPYRIMIDIN-5-YL)-1-PENTYL-PHOSPHONIC ACID, LUMAZINE SYNTHASE | | Authors: | Meining, W, Mortl, S, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue.
J.Mol.Biol., 299, 2000
|
|
1AGB
 
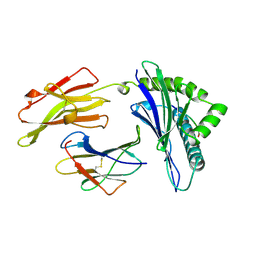 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGRKKYKL-3R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGRKKYKL - 3R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
4E1N
 
 | |
1EV4
 
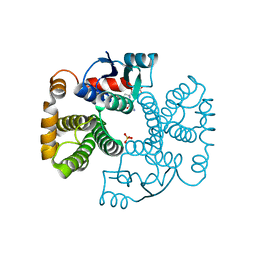 | | RAT GLUTATHIONE S-TRANSFERASE A1-1: MUTANT W21F/F220Y WITH GSO3 BOUND | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1-1, GLUTATHIONE SULFONIC ACID, SULFATE ION | | Authors: | Adman, E.T, Le Trong, I, Stenkamp, R.E, Nieslanik, B.S, Dietze, E.C, Tai, G, Ibarra, C, Atkins, W.M. | | Deposit date: | 2000-04-19 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Localization of the C-terminus of rat glutathione S-transferase A1-1: crystal structure of mutants W21F and W21F/F220Y.
Proteins, 42, 2001
|
|
4G4I
 
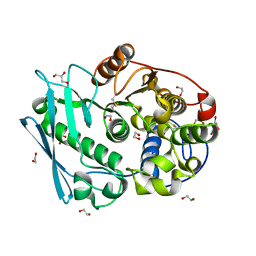 | | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile determined at 1.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-glucuronoyl methylesterase, GLYCEROL | | Authors: | Charvagi, M.D, Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2012-07-16 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3FFR
 
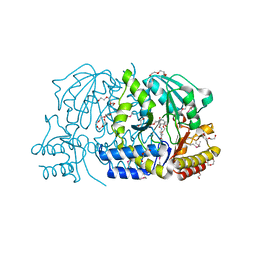 | |
4M80
 
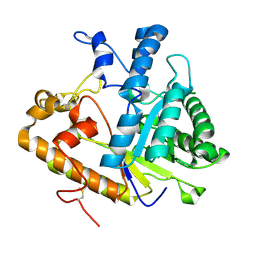 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution | | Descriptor: | EXO-1,3-BETA-GLUCANASE | | Authors: | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
4E1M
 
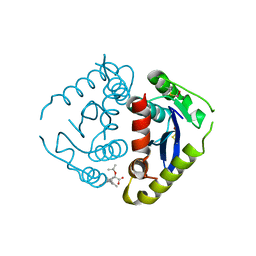 | |
1AAQ
 
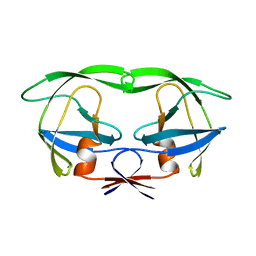 | |
3CT9
 
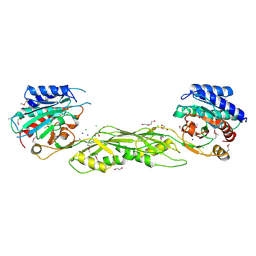 | |
4OFK
 
 | | Crystal Structure of SYG-2 D4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Ozkan, E, Borek, D, Otwinowski, Z, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OQ5
 
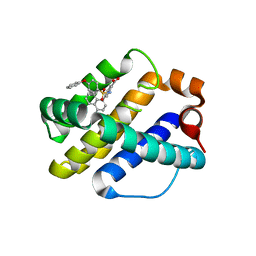 | | Crystal Structure of Human MCL-1 Bound to Inhibitor 4-(4-methylnaphthalen-1-yl)-2-{[(4-phenoxyphenyl)sulfonyl]amino}benzoic acid | | Descriptor: | 4-(4-methylnaphthalen-1-yl)-2-{[(4-phenoxyphenyl)sulfonyl]amino}benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Petros, A.M, Swann, S.L, Song, D, Swinger, K, Park, C, Zhang, H, Wendt, M.D, Kunzer, A.R, Souers, A.J, Sun, C. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Fragment-based discovery of potent inhibitors of the anti-apoptotic MCL-1 protein.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3TAW
 
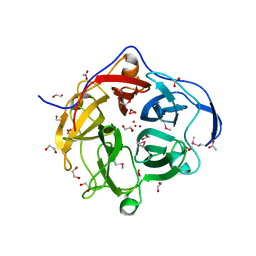 | |
3O0L
 
 | |
1R8L
 
 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis | | Descriptor: | CALCIUM ION, endo-beta-1,4-galactanase | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products
J.Mol.Biol., 341, 2004
|
|
6DFF
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef monomer | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Ren, X, Hurley, J.H. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
