3C7F
 
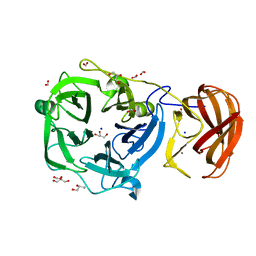 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C8T
 
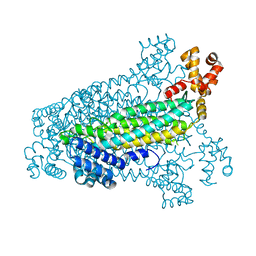 | | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1 | | Descriptor: | Fumarate lyase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-13 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1.
To be Published
|
|
3C8U
 
 | |
9EZB
 
 | |
3BL6
 
 | | Crystal structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Siu, K.K.W, Lee, J.E, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3BLI
 
 | |
8P8A
 
 | | Structure of 5D3-Fab and nanobody(Nb17)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
3BLN
 
 | |
9FBG
 
 | |
6CG2
 
 | | Crystal Structure of KDM4A with Compound 8 | | Descriptor: | 2-[5-(3-hydroxyphenyl)-1H-pyrazol-1-yl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J, Nie, Z. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based design and discovery of potent and selective KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3BKJ
 
 | | Crystal structure of Fab wo2 bound to the n terminal domain of amyloid beta peptide (1-16) | | Descriptor: | Amyloid Beta Peptide, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3BKY
 
 | | Crystal Structure of Chimeric Antibody C2H7 Fab in complex with a CD20 Peptide | | Descriptor: | B-lymphocyte antigen CD20, the Fab fragment of chimeric 2H7, heavy chain, ... | | Authors: | Du, J, Zhong, C, Ding, J. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of chimeric antibody C2H7 Fab in complex with a CD20 peptide
Mol.Immunol., 45, 2008
|
|
7XWW
 
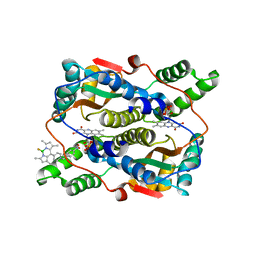 | | Crystal structure of NTR in complex with BN-XB | | Descriptor: | 2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-8-[1-[(4-nitrophenyl)methyl]pyridin-1-ium-4-yl]-3-aza-1-azonia-2-boranuidatricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaene, Dihydropteridine reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Chen, X, Chen, J, Li, J.L. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of NTR in complex with BN-XB
To Be Published
|
|
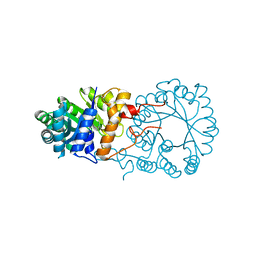
3BLE
 
 | |
7XZ3
 
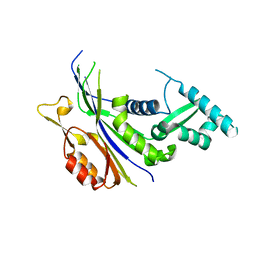 | | Crystal structure of the Type I-B CRISPR-associated protein, Csh2 from Thermobaculum terrenum | | Descriptor: | CRISPR-associated protein, Csh2 family | | Authors: | Seo, P.W, Gu, D.H, Park, S.Y, Kim, J.S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural characterization of the type I-B CRISPR Cas7 from Thermobaculum terrenum.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
7XRX
 
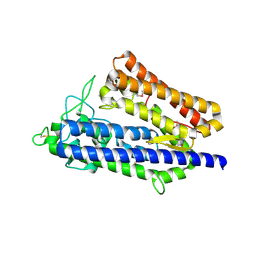 | | insulin-cleaving membrane protease-ICMP | | Descriptor: | CALCIUM ION, Insulin-cleaving metalloproteinase outer membrane protein | | Authors: | Wang, J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Crystallization and X-ray diffraction analysis of native and selenomethionine-substituted ICMP from P. aeruginosa
To Be Published
|
|
7XPQ
 
 | |
3BQ7
 
 | |
3BOR
 
 | | Crystal structure of the DEADc domain of human translation initiation factor 4A-2 | | Descriptor: | Human initiation factor 4A-II | | Authors: | Dimov, S, Hong, B, Tempel, W, MacKenzie, F, Karlberg, T, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3BQH
 
 | |
7XR8
 
 | |
7XRO
 
 | |
7XT1
 
 | |
7XQC
 
 | |
7XS0
 
 | | Trimer structure of HtrA from Helicobacter pylori bound with a tripeptide | | Descriptor: | Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK | | Authors: | Cui, L, Liu, W. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Int.J.Biol.Macromol., 243, 2023
|
|
