8AQ1
 
 | | small molecule stabilizer for ERalpha and 14-3-3 (1083743) | | Descriptor: | (2S,6R)-N-[[7-(2-chloranylethanoyl)-7-azaspiro[3.5]nonan-2-yl]methyl]-4-[(4-chlorophenyl)amino]-2,6-dimethyl-oxane-4-carboxamide, 14-3-3 protein sigma, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-11 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
6Q2T
 
 | | Human sterol 14a-demethylase (CYP51) in complex with the functionally irreversible inhibitor (R)-N-(1-(3-chloro-4'-fluoro-[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Lanosterol 14-alpha demethylase, N-[(1R)-1-(3-chloro-4'-fluoro[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, ... | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Validation of Human Sterol 14 alpha-Demethylase (CYP51) Druggability: Structure-Guided Design, Synthesis, and Evaluation of Stoichiometric, Functionally Irreversible Inhibitors.
J.Med.Chem., 62, 2019
|
|
7DVN
 
 | | Crystal structure of a MarR family protein in complex with a lipid-like effector molecule from the psychrophilic bacterium Paenisporosarcina sp. TG-14 | | Descriptor: | MarR family transcriptional regulator, PALMITIC ACID | | Authors: | Lee, C.W, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a MarR family protein from the psychrophilic bacterium Paenisporosarcina sp. TG-14 in complex with a lipid-like molecule.
Iucrj, 8, 2021
|
|
4DHS
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3,5-dichlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 PROTEIN SIGMA, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHR
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(2-chlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHO
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3-methoxyphenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHU
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(2,3-dichlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 PROTEIN SIGMA, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHQ
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3-chlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
6YIC
 
 | |
6YIA
 
 | |
6YIB
 
 | |
6MRW
 
 | | 14-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
5WFX
 
 | |
2FS3
 
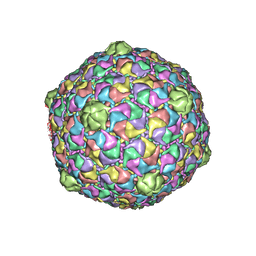 | | Bacteriophage HK97 K169Y Head I | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
4GNT
 
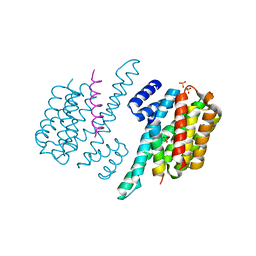 | | Complex of ChREBP and 14-3-3beta | | Descriptor: | 14-3-3 protein beta/alpha, Carbohydrate-responsive element-binding protein, SULFATE ION | | Authors: | Zhang, H, Huang, N. | | Deposit date: | 2012-08-17 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of Carbohydrate Response Element Binding Protein (ChREBP) in complex with 14-3-3b
To be Published
|
|
4QLI
 
 | |
3M50
 
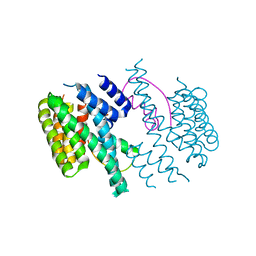 | | Structure of the 14-3-3/PMA2 complex stabilized by Epibestatin | | Descriptor: | 14-3-3-like protein C, N-[(2R,3R)-3-amino-2-hydroxy-4-phenylbutanoyl]-L-leucine, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Ottmann, C, Rose, R, Waldmann, H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3M51
 
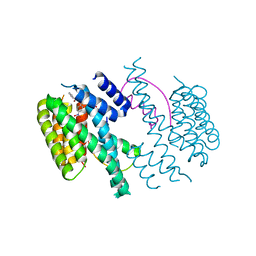 | | Structure of the 14-3-3/PMA2 complex stabilized by Pyrrolidone1 | | Descriptor: | 14-3-3-like protein C, 2-hydroxy-5-[(5S)-3-hydroxy-5-(4-nitrophenyl)-2-oxo-4-(phenylcarbonyl)-2,5-dihydro-1H-pyrrol-1-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Ottmann, C, Rose, R, Waldmann, H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
4WV1
 
 | |
5WFU
 
 | |
8XI3
 
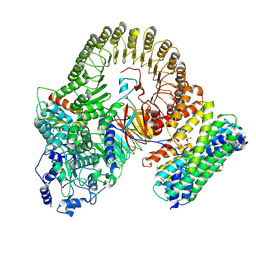 | | Structure of mouse SCMC-14-3-3gama complex | | Descriptor: | 14-3-3 protein gamma, NACHT, LRR and PYD domains-containing protein 5, ... | | Authors: | Chi, P, Han, Z, Jiao, H, Li, J, Wang, X, Hu, H, Deng, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of mouse SCMC-14-3-3gama complex
To Be Published
|
|
5XY9
 
 | | Structure of the MST4 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2017-07-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
5LVZ
 
 | | Crystal structure of yeast 14-3-3 protein from Lachancea thermotolerans | | Descriptor: | KLTH0G14146p | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of a yeast 14-3-3 protein from Lachancea thermotolerans in the unliganded form and bound to a human lipid kinase PI4KB-derived peptide reveal high evolutionary conservation.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6EJL
 
 | |
3U9X
 
 | | Covalent attachment of pyridoxal-phosphate derivatives to 14-3-3 proteins | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-09 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent attachment of pyridoxal-phosphate derivatives to 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
