7I1Y
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with 3632-JP-070-004 | | Descriptor: | (1s,3s)-3-amino-N-[4-(hydroxymethyl)-2-methylquinolin-8-yl]cyclobutane-1-carboxamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Balaji, G, Phelps, J, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2025-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7I1W
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with 3632-JP-070-010 | | Descriptor: | (1r,4r)-4-amino-N-[4-(hydroxymethyl)-2-methylquinolin-8-yl]cyclohexane-1-carboxamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Balaji, G, Phelps, J, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2025-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4YQ2
 
 | |
9GE2
 
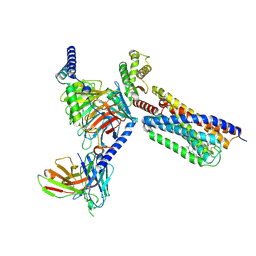 | | Structure of GPR55 in complex with G13 and synthetic agonist ML184 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2,3-dimethylphenyl)piperazin-1-yl]carbonyl-N,N-dimethyl-4-pyrrolidin-1-yl-benzenesulfonamide, CHOLESTEROL, ... | | Authors: | Claff, T, Ebenhoch, R, Weichert, D. | | Deposit date: | 2024-08-06 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural basis for lipid-mediated activation of G protein-coupled receptor GPR55.
Nat Commun, 16, 2025
|
|
6QTZ
 
 | |
4YRG
 
 | |
8TRL
 
 | | T cell recognition of citrullinated alpha-enolase peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
7YKN
 
 | | Crystal structure of (6-4) photolyase from Vibrio cholerae | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cakilkaya, B, Kavakli, I.H, DeMirci, H. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Vibrio cholerae (6-4) photolyase reveals interactions with cofactors and a DNA-binding region.
J.Biol.Chem., 299, 2023
|
|
1H8C
 
 | | UBX domain from human faf1 | | Descriptor: | FAS-ASSOCIATED FACTOR 1 | | Authors: | Bycroft, M.M. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Ubx Domain: A Widespread Ubiquitin-Like Module
J.Mol.Biol., 307, 2001
|
|
6F56
 
 | |
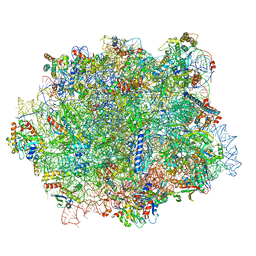
8B2L
 
 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
6GZ3
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-1 (TI-POST-1) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
6QTP
 
 | | 2.37A structure of gepotidacin with S.aureus DNA gyrase and uncleaved DNA | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*AP*GP*CP*TP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D. | | Deposit date: | 2019-02-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase.
Acs Infect Dis., 5, 2019
|
|
4YRZ
 
 | | Crystal Structure of LuxP In Complex With L-xylulofuranose-1,2-borate | | Descriptor: | Autoinducer 2-binding periplasmic protein LuxP, L-xylulofuranose-1,2-borate | | Authors: | McDonough, M.A, Sattin, S, Gardner, P.M, Liu, C, Davis, B.G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.571 Å) | | Cite: | Discovery of Glycomimetic Agonists from a Protocell Metabolism, Proto-natural Product Libraries
To Be Published
|
|
3IPY
 
 | | X-Ray structure of Human Deoxycytidine Kinase in complex with an inhibitor | | Descriptor: | 4-(1-benzothiophen-2-yl)-6-[4-(2-oxo-2-pyrrolidin-1-ylethyl)piperazin-1-yl]pyrimidine, D-MALATE, Deoxycytidine kinase | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M, Hoffman, I, Stouch, T.R, Carson, K.G. | | Deposit date: | 2009-08-18 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Lead optimization and structure-based design of potent and bioavailable deoxycytidine kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2YOB
 
 | | High resolution AGXT_M structure | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fabelo-Rosa, I, Mesa-Torres, N, Riverol, D, Yunta, C, Albert, A, Salido, E, Pey, A.L. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Role of Protein Denaturation Energetics and Molecular Chaperones in the Aggregation and Mistargeting of Mutants Causing Primary Hyperoxaluria Type I
Plos One, 8, 2013
|
|
5OUM
 
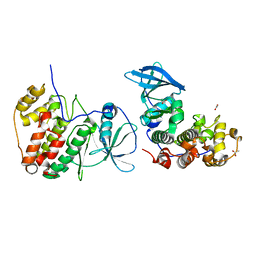 | | The crystal structure of CK2alpha in complex with compound 21 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, ~{N}-[(3-chloranyl-4-phenyl-phenyl)methyl]-2-(1~{H}-imidazol-2-yl)ethanamine | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-24 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
3DYS
 
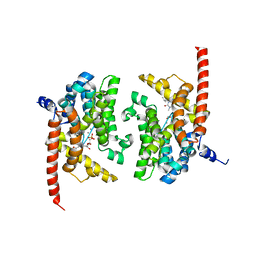 | | human phosphodiestrase-5'GMP complex (EP), produced by soaking with 20mM cGMP+20 mM MnCl2+20 mM MgCl2 for 2 hours, and flash-cooled to liquid nitrogen temperature when substrate was still abudant. | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, FORMIC ACID, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Liu, S, Mansour, M.N, Dillman, K, Perez, J, Danley, D, Menniti, F. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the catalytic mechanism of human phosphodiesterase 9.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4K1P
 
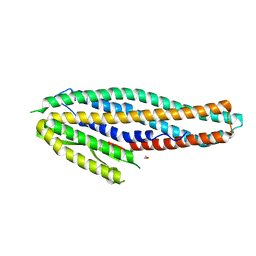 | | Structure of the NheA component of the Nhe toxin from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, NheA, SULFATE ION | | Authors: | Ganash, M, Phung, D, Artymiuk, P.J. | | Deposit date: | 2013-04-05 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the NheA Component of the Nhe Toxin from Bacillus cereus: Implications for Function.
Plos One, 8, 2013
|
|
5UE4
 
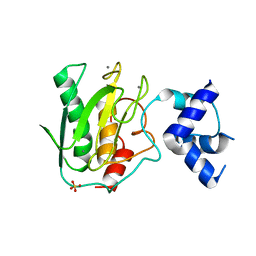 | | proMMP-9desFnII complexed to JNJ0966 INHIBITOR | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, SULFATE ION, ... | | Authors: | Alexander, R.S, Spurlino, J, Milligan, C. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
2WKS
 
 | | Structure of Helicobacter pylori Type II Dehydroquinase with a new carbasugar-thiophene inhibitor. | | Descriptor: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Guardado-Calvo, P, Llamas-Saiz, A.L, Prazeres, V.F.V, Tizon, L, Castedo, L, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues.
J. Med. Chem., 53, 2010
|
|
7WFC
 
 | | X-ray structure of HKU1-PLP2(Cys109Ser) catalytic mutant in complex with free ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, 60S ribosomal protein L40, Papain-like protease, ... | | Authors: | Xiong, Y.X, Fu, Z.Y, Huang, H. | | Deposit date: | 2021-12-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The substrate selectivity of papain-like proteases from human-infecting coronaviruses correlates with innate immune suppression.
Sci.Signal., 16, 2023
|
|
4K6I
 
 | | Crystal structure of human retinoid X receptor alpha-ligand binding domain complex with Targretin and the coactivator peptide GRIP-1 | | Descriptor: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2013-04-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining the Communication between Agonist and Coactivator Binding in the Retinoid X Receptor alpha Ligand Binding Domain.
J.Biol.Chem., 289, 2014
|
|
9K0J
 
 | |
1HDK
 
 | | Charcot-Leyden Crystal Protein - pCMBS Complex | | Descriptor: | EOSINOPHIL LYSOPHOSPHOLIPASE, PARA-MERCURY-BENZENESULFONIC ACID | | Authors: | Ackerman, S.J, Savage, M.P, Liu, L, Leonidas, D.D, Kwatia, M.A, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden Crystal Protein (Galectin-10) is not a Dual Function Galectin with Lysophospholipase Activity But Binds a Lysophospholipase Inhibitor in a Novel Structural Fashion.
J.Biol.Chem., 277, 2002
|
|
