5DV8
 
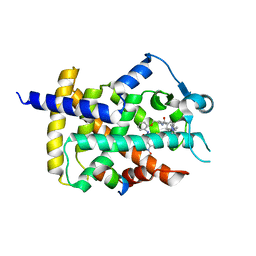 | | Human PPARgamma ligand binding dmain complexed with SB1451 in a covalent bonded form | | Descriptor: | N-[2-({2-[({4-[(4-methylpiperazin-1-yl)methyl]benzoyl}amino)methyl]benzyl}oxy)phenyl]-3-nitrobenzamide, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y. | | Deposit date: | 2015-09-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Human PPARgamma ligand binding dmain complexed with SB1451 in a covalent bonded form
To Be Published
|
|
3ALC
 
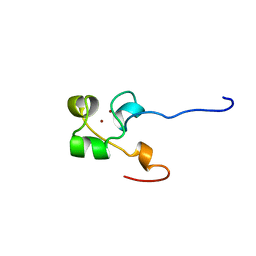 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-03-11 | | Release date: | 2000-05-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
2Y7C
 
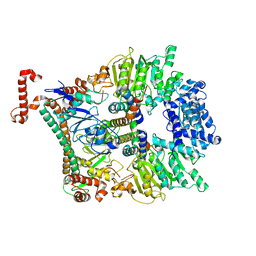 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
5DZ8
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA variable (V) domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BspA (BspA_V), ... | | Authors: | Rego, S, Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
6B4W
 
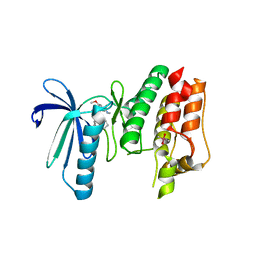 | | TTK in Complex with Inhibitor | | Descriptor: | 4-{[4-(cyclopentyloxy)-5-(2-methyl-1,3-benzoxazol-6-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}-3-methoxy-N-methylbenzamide, CACODYLATE ION, Dual specificity protein kinase TTK | | Authors: | Delker, S, Chamberlain, P.P. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery of a Dual TTK Protein Kinase/CDC2-Like Kinase (CLK2) Inhibitor for the Treatment of Triple Negative Breast Cancer Initiated from a Phenotypic Screen.
J. Med. Chem., 60, 2017
|
|
5DX1
 
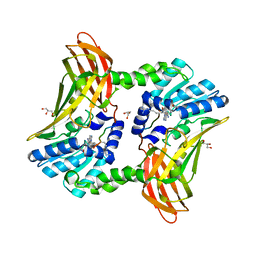 | |
1LRI
 
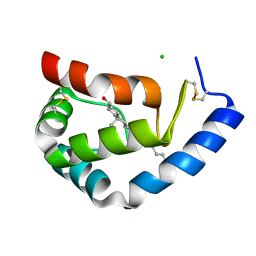 | | BETA-CRYPTOGEIN-CHOLESTEROL COMPLEX | | Descriptor: | Beta-elicitin cryptogein, CHLORIDE ION, CHOLESTEROL | | Authors: | Lascombe, M.-B, Ponchet, M, Venard, P, Milat, M.-L, Blein, J.-P, Prange, T. | | Deposit date: | 2002-05-15 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The 1.45 A resolution structure of the cryptogein-cholesterol complex: a close-up view of a sterol carrier protein (SCP) active site.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5HCT
 
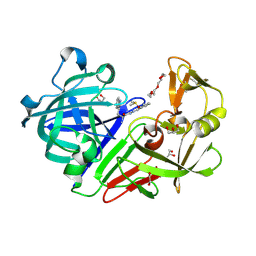 | | Endothiapepsin in complex with biacylhydrazone | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-N'-{3-[(E)-{2-[(2S)-2-amino-3-(1H-indol-3-yl)propanoyl]hydrazinylidene}methyl]benzylidene}-3-(1H-indol-2-yl)propanehydrazide (non-preferred name), ACETATE ION, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2016-01-04 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fragment Linking and Optimization of Inhibitors of the Aspartic Protease Endothiapepsin: Fragment-Based Drug Design Facilitated by Dynamic Combinatorial Chemistry.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3VIJ
 
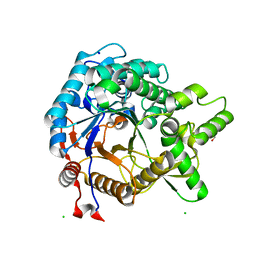 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with glucose | | Descriptor: | Beta-glucosidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1XCS
 
 | | structure of oligonucleotide/drug complex | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-[(5-(ACETYLAMINO)-6-{[(1S,4R)-8-AMINO-4-[((2R)-6-AMINO-2-{2-[(1S)-5-AMINO-1-FORMYLPENTYL]HYDRAZINO}HEXANOYL)AMINO]-1-(4-AMINOBUTYL)-2,3-DIOXOOCTYL]AMINO}-6-OXOHEXYL)AMINO]-6-CHLORO-2-METHOXYACRIDINIUM, BARIUM ION, ... | | Authors: | Valls, N, Steiner, R.A, Wright, G, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Variable role of ions in two drug intercalation complexes of DNA
J.Biol.Inorg.Chem., 10, 2005
|
|
1IJR
 
 | | Crystal structure of LCK SH2 complexed with nonpeptide phosphotyrosine mimetic | | Descriptor: | (4-{2-ACETYLAMINO-2-[1-(3-CARBAMOYL-4-CYCLOHEXYLMETHOXY-PHENYL)-ETHYLCARBAMOYL}-ETHYL}-2-PHOSPHONO-PHENOXY)-ACETIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Kawahata, N.H, Yang, M.H, Luke, G.P, Shakespeare, W.C, Sundaramoorthi, R. | | Deposit date: | 2001-04-27 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel phosphotyrosine mimetic 4'-carboxymethyloxy-3'-phosphonophenylalanine (Cpp): exploitation in the design of nonpeptide inhibitors of pp60(Src) SH2 domain.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
3NP2
 
 | | Crystal Structure of Pd(allyl)/apo-E45C/C48A-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Wang, Z, Ueno, T, Abe, S, Takezawa, Y, Aoyagi, H, Hikage, T, Watanabe, Y, Kitagawa, S. | | Deposit date: | 2010-06-27 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Definite coordination arrangement of organometallic palladium complexes accumulated on the designed interior surface of apo-ferritin.
Chem.Commun.(Camb.), 47, 2011
|
|
4OIN
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
5CHM
 
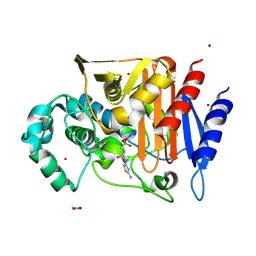 | | CRYSTAL STRUCTURE OF Fox-4 cephamycinase complexed with ceftazidime BATSI (LP06) | | Descriptor: | ACETATE ION, Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, ... | | Authors: | Malashkevich, V.N, Toro, R, Lefurgy, S, Almo, S.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of FOX-4 Cephamycinase in Complex with Transition-State Analog Inhibitors.
Biomolecules, 10, 2020
|
|
5SP1
 
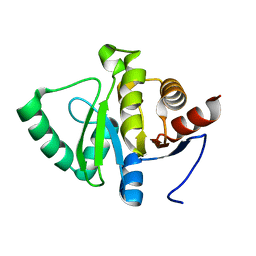 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC001472868186 | | Descriptor: | 3-{[methyl(pyrido[2,3-b]pyrazin-6-yl)amino]methyl}[1,2,4]triazolo[4,3-a]pyrazin-8(7H)-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SP6
 
 | |
4OIO
 
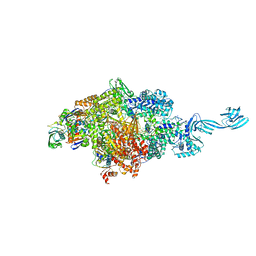 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
1PB7
 
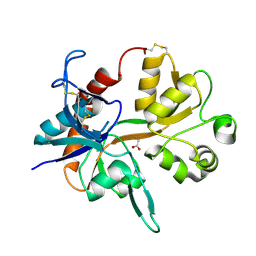 | |
3GE7
 
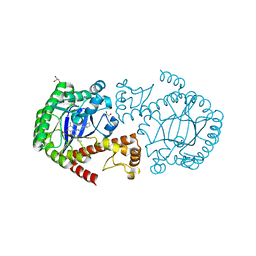 | | tRNA-guanine transglycosylase in complex with 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2009-02-25 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
2Y1S
 
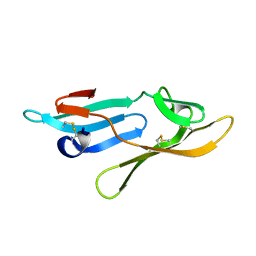 | | Microvirin lectin | | Descriptor: | Mannan-binding lectin | | Authors: | Bewley, C.A, Hussan, S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monovalent lectin microvirin in complex with Man(alpha)(1-2)Man provides a basis for anti-HIV activity with low toxicity.
J. Biol. Chem., 286, 2011
|
|
2Y4J
 
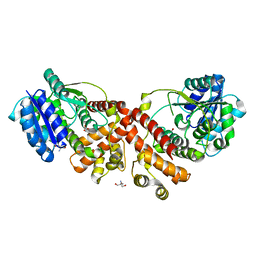 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH LACTATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LACTIC ACID, MANNOSYLGLYCERATE SYNTHASE, ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
5C3K
 
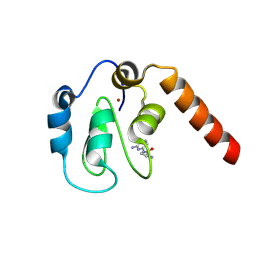 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 4 | | Descriptor: | (2S)-1-[(6-aminopyridin-2-yl)amino]-1-oxopropan-2-aminium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6FG3
 
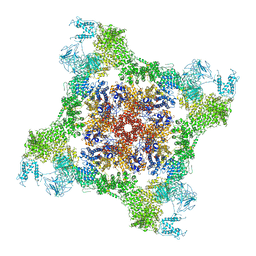 | |
5C7B
 
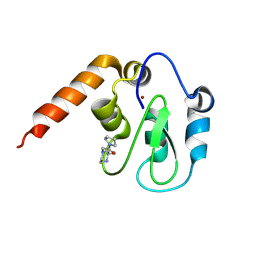 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 5 | | Descriptor: | (2R)-2-methyl-4-[2-oxo-2-(pyrrolidin-1-yl)ethyl]piperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6FKY
 
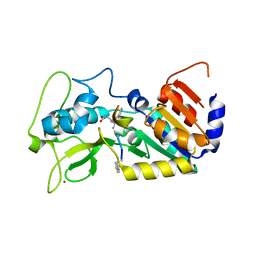 | |
