7XFW
 
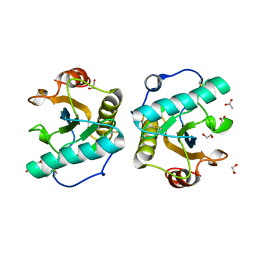 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.07 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the complex of camel peptidoglycan recognition protein-S with hexanoic acid reveals novel features of the versatile ligand-binding site at the dimeric interface.
Biochim Biophys Acta Proteins Proteom, 1871, 2022
|
|
7XFX
 
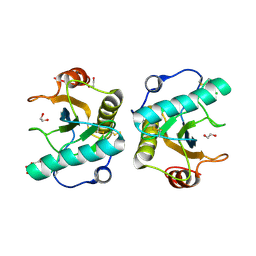 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution.
To Be Published
|
|
7XFY
 
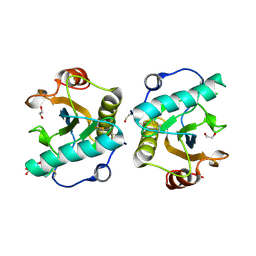 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution.
To Be Published
|
|
2PVE
 
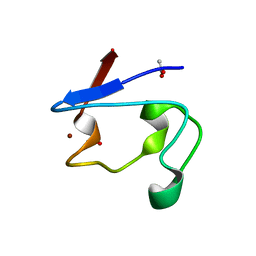 | | NMR and X-ray Analysis of Structural Additivity in Metal Binding Site-Swapped Hybrids of Rubredoxin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Rubredoxin, ... | | Authors: | LeMaster, D.M, Anderson, J.S, Wang, L, Guo, Y, Li, H, Hernandez, G. | | Deposit date: | 2007-05-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.79 Å) | | Cite: | NMR and X-ray analysis of structural additivity in metal binding site-swapped hybrids of rubredoxin.
Bmc Struct.Biol., 7, 2007
|
|
7F03
 
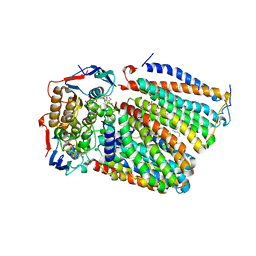 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F02
 
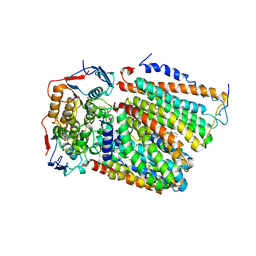 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
1C44
 
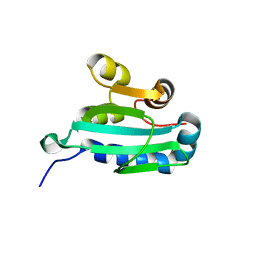 | |
5DSO
 
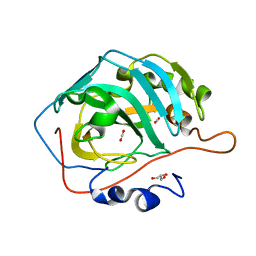 | |
5HVS
 
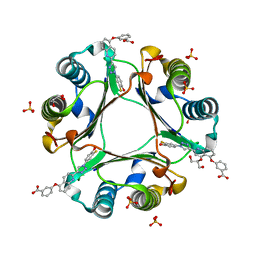 | | Crystal Structure of Macrophage Migration Inhibitory Factor (MIF) with a Biaryltriazole Inhibitor (3i-305) | | Descriptor: | 3-({2-[1-(3-fluoro-4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]quinolin-5-yl}oxy)benzoic acid, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2016-01-28 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Fluorescence Polarization Assay for Binding to Macrophage Migration Inhibitory Factor and Crystal Structures for Complexes of Two Potent Inhibitors.
J.Am.Chem.Soc., 138, 2016
|
|
6FJQ
 
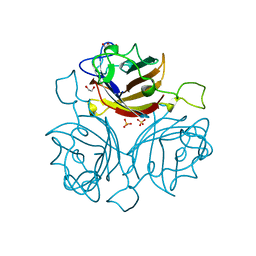 | | Adenovirus species 48, fiber knob protein | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Nat Commun, 10, 2019
|
|
5HWY
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 10 mM Na+ and zero Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ACETATE ION, PENTADECANE, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger
Nat.Struct.Mol.Biol., 23, 2016
|
|
4IDE
 
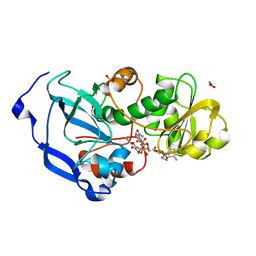 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADP+ and EDHMF | | Descriptor: | (2E)-2-ethylidene-4-hydroxy-5-methylfuran-3(2H)-one, 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
7VQG
 
 | | The X-ray structure of human neuroglobin A15C mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Neuroglobin, ... | | Authors: | Lin, Y.W. | | Deposit date: | 2021-10-19 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The X-ray structure of human neuroglobin A15C mutant
To Be Published
|
|
4G3M
 
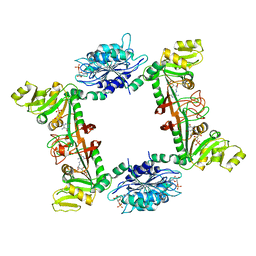 | | Complex Structure of Bacillus subtilis RibG: The Deamination Process in Riboflavin Biosynthesis | | Descriptor: | N-(5-amino-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)-5-O-phosphono-beta-D-ribofuranosylamine, Riboflavin biosynthesis protein RibD, ZINC ION, ... | | Authors: | Chen, S.C, Shen, C.Y, Yen, T.M, Yu, H.C, Chang, T.H, Lai, W.L, Liaw, S.H. | | Deposit date: | 2012-07-15 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Evolution of vitamin B(2) biosynthesis: eubacterial RibG and fungal Rib2 deaminases.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2Q3W
 
 | | Ensemble refinement of the protein crystal structure of the cys84ala cys85ala double mutant of the [2Fe-2S] ferredoxin subunit of toluene-4-monooxygenase from Pseudomonas mendocina KR1 | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
4IDD
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADPH and EHMF | | Descriptor: | (2R)-2-ethyl-4-hydroxy-5-methylfuran-3(2H)-one, 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
6J5I
 
 | | Cryo-EM structure of the mammalian DP-state ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit epsilon, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
7K0A
 
 | | Puromycin N-acetyltransferase in complex with acetylated puromycin and CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Caputo, A.T, Newman, J, Adams, T.E. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
3UN8
 
 | | Yeast 20S proteasome in complex with PR-957 (epoxide) | | Descriptor: | 2-(acetylamino)-4,5-anhydro-1,2-dideoxy-4-methyl-1-phenyl-D-xylitol, Proteasome component C1, Proteasome component C11, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
1IWN
 
 | | Crystal Structure of the Outer Membrane Lipoprotein Receptor LolB Complexed with PEGMME2000 | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Outer Membrane Lipoprotein LolB, SULFATE ION | | Authors: | Takeda, K, Miyatake, H, Yokota, N, Matsuyama, S, Tokuda, H, Miki, K. | | Deposit date: | 2002-05-17 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of bacterial lipoprotein localization factors, LolA and LolB.
Embo J., 22, 2003
|
|
6CYQ
 
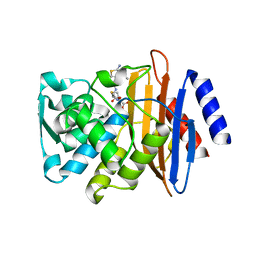 | | Crystal structure of CTX-M-14 S70G/N106S beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, GLYCEROL | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
1OOT
 
 | |
6CZV
 
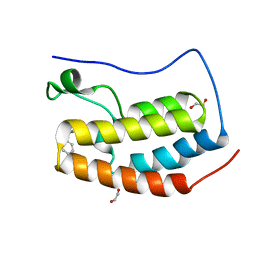 | | BRD4(BD1) complexed with 2759 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-5-(3,5-dimethyl-1,2-oxazol-4-yl)pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
7JQD
 
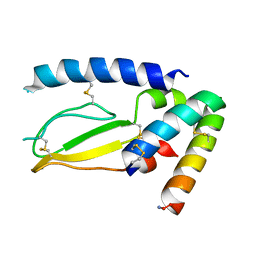 | | Crystal Structure of PAC1r in complex with peptide antagonist | | Descriptor: | Peptide-43, Pituitary adenylate cyclase-activating polypeptide type I receptor | | Authors: | Piper, D.E, Hu, E, Fang-Tsao, H. | | Deposit date: | 2020-08-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Selective Pituitary Adenylate Cyclase 1 Receptor (PAC1R) Antagonist Peptides Potent in a Maxadilan/PACAP38-Induced Increase in Blood Flow Pharmacodynamic Model.
J.Med.Chem., 64, 2021
|
|
2YSA
 
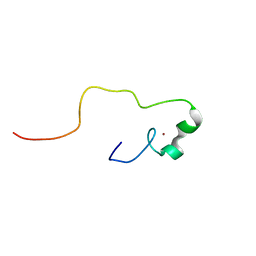 | | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1) | | Descriptor: | Retinoblastoma-binding protein 6, ZINC ION | | Authors: | Ohnishi, S, Sato, M, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1)
To be Published
|
|
