7R8Q
 
 | | Closed form of SAOUHSC_02373 in complex with ADP, citrate, Mg2+ and Na+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-grasp domain-containing protein, CITRIC ACID, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-06-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of an ʟ-amino acid ligase implicated in Staphylococcal sulfur amino acid metabolism.
J.Biol.Chem., 298, 2022
|
|
6VAV
 
 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-succinamoyl derivative (diNGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC3
 
 | | Peanut lectin complexed with S-beta-D-thiogalactopyranosyl 6-deoxy-6-S-propynyl-beta-D-glucopyranoside (STG) | | Descriptor: | 6-S-(prop-2-yn-1-yl)-6-thio-beta-D-glucopyranosyl 1-thio-beta-D-galactopyranoside, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4TT2
 
 | | Crystal structure of ATAD2A bromodomain complexed with H4(1-20)K5Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4K5Ac | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
3UWU
 
 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with glycerol-3-phosphate | | Descriptor: | CITRIC ACID, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
8FQ9
 
 | |
4TU6
 
 | | Crystal structure of apo ATAD2A bromodomain with N1064 alternate conformation | | Descriptor: | ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU4
 
 | | Crystal structure of ATAD2A bromodomain complexed with 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoicacid | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoic acid, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
7UWM
 
 | | Structure of the IL-17A-IL-17RA binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
8I01
 
 | | Crystal structure of Escherichia coli glyoxylate carboligase | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8I05
 
 | | Crystal structure of Escherichia coli glyoxylate carboligase double mutant | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8I07
 
 | |
8I08
 
 | | Crystal structure of Escherichia coli glyoxylate carboligase quadruple mutant | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
7UWN
 
 | | Structure of the IL-17A-IL-17RA-IL-17RC ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
4TT6
 
 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
5OBC
 
 | | X-ray structure of the adduct formed upon reaction of ribonuclease A with the compound fac-[RuII(CO)3Cl2(N3-IM), IM=imidazole | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic, pentakis(oxidaniumyl)-(oxidaniumylidynemethyl)ruthenium, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
3H6V
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | Descriptor: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
6ASI
 
 | | E. coli phosphoenolpyruvate carboxykinase G209S mutant bound to methanesulfonate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6AT2
 
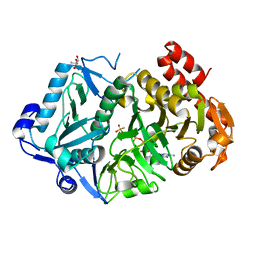 | | E. coli phosphoenolpyruvate carboxykinase G209N mutant bound to thiosulfate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-27 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
2RKM
 
 | | STRUCTURE OF OPPA COMPLEXED WITH LYS-LYS | | Descriptor: | LYSINE, OLIGO-PEPTIDE BINDING PROTEIN, URANYL (VI) ION | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
4AVD
 
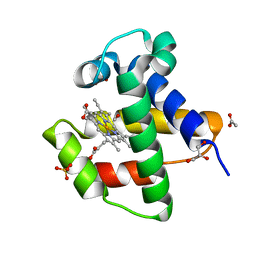 | | C.lacteus nerve Hb in complex with CO | | Descriptor: | ACETATE ION, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Germani, F, Pesce, A, Venturini, A, Moens, L, Bolognesi, M, Dewilde, S, Nardini, M. | | Deposit date: | 2012-05-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Crystal Structures of the Cerebratulus Lacteus Mini-Hb in the Unligated and Carbomonoxy States.
Int.J.Mol.Sci., 13, 2012
|
|
4AVE
 
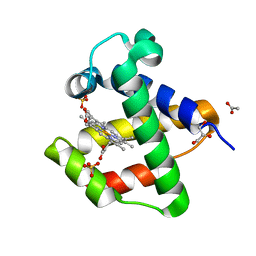 | | C.lacteus nerve Hb in the deoxy form | | Descriptor: | ACETATE ION, GLYCEROL, NEURAL HEMOGLOBIN, ... | | Authors: | Germani, F, Pesce, A, Venturini, A, Moens, L, Bolognesi, M, Dewilde, S, Nardini, M. | | Deposit date: | 2012-05-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Crystal Structures of the Cerebratulus Lacteus Mini-Hb in the Unligated and Carbomonoxy States.
Int.J.Mol.Sci., 13, 2012
|
|
4H98
 
 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-{3-[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine (UCP1018) | | Descriptor: | 5-{3-[3-(1,3-benzodioxol-5-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine, Dihydrofolate Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-09-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6VAW
 
 | | Peanut lectin complexed with N-beta-D-galactopyranosyl-L-succinamoyl derivative (NGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
