1GP2
 
 | |
4HE1
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase Q32R mutant complex with fructose-6-phosphate and phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
3K11
 
 | |
4M0N
 
 | |
2UY4
 
 | | ScCTS1_acetazolamide crystal structure | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, ENDOCHITINASE | | Authors: | Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Saccharomyces Cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors.
Chem.Biol., 14, 2007
|
|
5ZGE
 
 | | Crystal structure of NDM-1 at pH5.5 (Bis-Tris) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGR
 
 | | Crystal structure of NDM-1 at pH7.3 (HEPES) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
7LY9
 
 | |
4ISO
 
 | | Crystal Structure of Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, GLYCEROL, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
1FTA
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE, 1-PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH THE ALLOSTERIC INHIBITOR AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Lipscomb, W.N. | | Deposit date: | 1993-09-27 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric site of human liver fructose-1,6-bisphosphatase. Analysis of six AMP site mutants based on the crystal structure.
J.Biol.Chem., 269, 1994
|
|
4A9K
 
 | | BROMODOMAIN OF HUMAN CREBBP WITH N-(4-hydroxyphenyl)acetamide | | Descriptor: | 1,2-ETHANEDIOL, CREB-BINDING PROTEIN, N-(4-HYDROXYPHENYL)ACETAMIDE (TYLENOL), ... | | Authors: | Chung, C.W, Bamborough, P. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-Based Discovery of Bromodomain Inhibitors Part 1: Inhibitor Binding Modes and Implications for Lead Discovery.
J.Med.Chem., 55, 2012
|
|
3KGY
 
 | |
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
4AA5
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 33 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-CYCLOPROPYL-4-METHYL-3-[6-(4-METHYLPIPERAZIN-1-YL)-4-OXIDANYLIDENE-QUINAZOLIN-3-YL]BENZAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2UY3
 
 | |
7UGN
 
 | |
6PNO
 
 | |
3NDM
 
 | |
4GW6
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](tert-butoxy)ethanoic acid, ARSENIC, Gag-Pol polyprotein | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The A128T Resistance Mutation Reveals Aberrant Protein Multimerization as the Primary Mechanism of Action of Allosteric HIV-1 Integrase Inhibitors.
J.Biol.Chem., 288, 2013
|
|
7UGP
 
 | |
1GT7
 
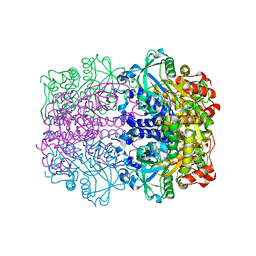 | | L-rhamnulose-1-phosphate aldolase from Escherichia coli | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, RHAMNULOSE-1-PHOSPHATE ALDOLASE, ZINC ION | | Authors: | Kroemer, M, Schulz, G.E. | | Deposit date: | 2002-01-14 | | Release date: | 2002-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of L-Rhamnulose-1-Phosphate Aldolase (Class II) Solved by Low-Resolution Sir Phasing and 20-Fold Ncs Averaging
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5VPF
 
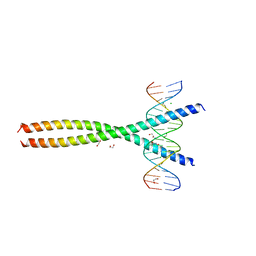 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Rudenko, G, Machius, M. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
1VMB
 
 | |
3LUL
 
 | |
3KLI
 
 | |
