7AHN
 
 | | Cryo-EM structure of F-actin stabilized by cis-optoJASP-8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Raunser, S. | | Deposit date: | 2020-09-24 | | Release date: | 2021-01-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Resolves Molecular Recognition Of An Optojasp Photoswitch Bound To Actin Filaments In Both Switch States.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B16
 
 | | TRPC4 in complex with inhibitor GFB-9289 | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 5-chloranyl-4-(4-cyclohexyl-3-oxidanylidene-piperazin-1-yl)-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B0S
 
 | | TRPC4 in complex with inhibitor GFB-8438 | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 5-chloranyl-4-[3-oxidanylidene-4-[[2-(trifluoromethyl)phenyl]methyl]piperazin-1-yl]-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B1G
 
 | | TRPC4 in complex with Calmodulin | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, CALCIUM ION, Calmodulin-1, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B0J
 
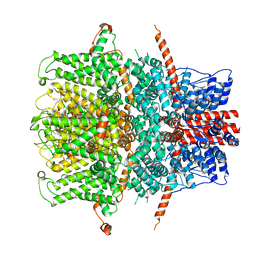 | | TRPC4 in LMNG detergent | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, Transient receptor potential cation channel subfamily c member 4a | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7AT8
 
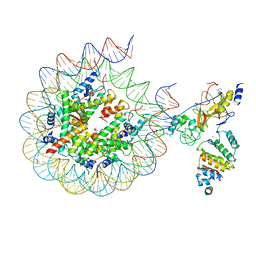 | | Histone H3 recognition by nucleosome-bound PRC2 subunit EZH2. | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | Authors: | Finogenova, K, Benda, C, Schaefer, I.B, Poepsel, S, Strauss, M, Mueller, J. | | Deposit date: | 2020-10-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Elife, 9, 2020
|
|
7B05
 
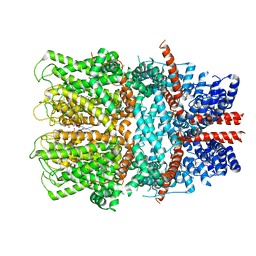 | | TRPC4 in complex with inhibitor GFB-8749 | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4-[4-[[4,4-bis(fluoranyl)cyclohexyl]methyl]-3-oxidanylidene-piperazin-1-yl]-5-chloranyl-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7AHQ
 
 | | Cryo-EM structure of F-actin stabilized by trans-optoJASP-8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Raunser, S. | | Deposit date: | 2020-09-25 | | Release date: | 2021-01-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Resolves Molecular Recognition Of An Optojasp Photoswitch Bound To Actin Filaments In Both Switch States.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ARV
 
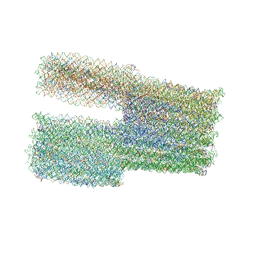 | | TwistTower_native-twist | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ARY
 
 | | Twist-Tower_twist-corrected-variant | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
1XA9
 
 | | Crystal structure of yellow fluorescent protein zFP538 K66M green mutant | | Descriptor: | BETA-MERCAPTOETHANOL, fluorescent protein FP538 | | Authors: | Remington, S.J, Wachter, R.M, Yarbrough, D.K, Branchaud, B, Anderson, D.C, Kallio, K, Lukyanov, K.A. | | Deposit date: | 2004-08-25 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore.
Biochemistry, 44, 2005
|
|
6ZYM
 
 | | Human C Complex Spliceosome - High-resolution CORE | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Corepressor interacting with RBPJ 1, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
7ADO
 
 | | Cryo-EM structure of human ER membrane protein complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7A5P
 
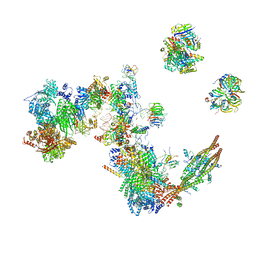 | | Human C Complex Spliceosome - Medium-resolution PERIPHERY | | Descriptor: | Cell division cycle 5-like protein, Crooked neck-like protein 1, Eukaryotic initiation factor 4A-III, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
7A8X
 
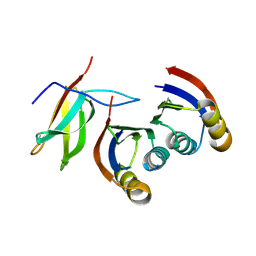 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with the HMA domain of Pikh-1 from rice (Oryza sativa) | | Descriptor: | AVR-Pik protein, NBS-LRR class disease resistance protein | | Authors: | Maidment, J.H.R, Xiao, G, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
7B2C
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum gassed with xenon | | Descriptor: | (2R)-2-[(2S)-2-[(2S)-2-oxidanylpropoxy]propoxy]propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
7B2H
 
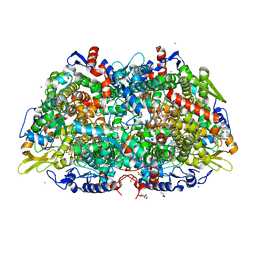 | | Crystal structure of the methyl-coenzyme M reductase from Methanothermobacter Marburgensis derivatized with xenon | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
7B1S
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum at 0.994-A resolution | | Descriptor: | (2S)-2-{[(2S)-2-{[(2S)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-25 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.992 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
7A6B
 
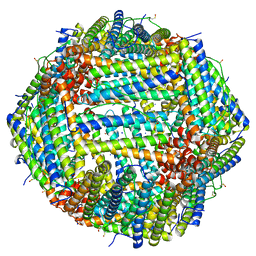 | | 1.33 A structure of human apoferritin obtained from Titan Mono- BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Chari, A, Stark, H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-09-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (1.33 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
4WP4
 
 | | Hev b 6.02 (hevein) extracted from surgical gloves | | Descriptor: | Pro-hevein, THIOUREA | | Authors: | Galicia, C, Rodriguez-Romero, A. | | Deposit date: | 2014-10-17 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Impact of the vulcanization process on the structural characteristics and IgE recognition of two allergens, Hev b 2 and Hev b 6.02, extracted from latex surgical gloves.
Mol.Immunol., 65, 2015
|
|
7ADP
 
 | | Cryo-EM structure of human ER membrane protein complex in GDN detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7AU6
 
 | |
7ATN
 
 | |
7ATE
 
 | | Cytochrome c oxidase structure in P-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-10-30 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cytochrome c oxidase structure in P-state
To Be Published
|
|
7AU3
 
 | | Cytochrome c oxidase structure in F-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cytochrome c oxidase structure in F-state
To Be Published
|
|
