2HXZ
 
 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor (Hexagonal spacegroup) | | Descriptor: | Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE, SULFATE ION | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
2I78
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPP IV) complexed with ABT-341, a cyclohexene-constrained phenethylamine inhibitor | | Descriptor: | (1S,6R)-3-{[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]CARBONYL}-6-(2,4,5-TRIFLUOROPHENYL)CYCLOHEX-3-EN-1-AMINE, Dipeptidyl peptidase IV | | Authors: | Longenecker, K.L, Pei, Z, Li, X. | | Deposit date: | 2006-08-30 | | Release date: | 2007-10-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Cyclohexene-constrained Phenethylamine ABT-341, a Highly Potent, Selective, Orally Bioavailable, Safe and Potential Next-generation Dipeptidyl Peptidase IV Inhibitor for the Treatment of Type 2 Diabetes
To be Published
|
|
6DEM
 
 | |
7OMF
 
 | | Structure of a minimal SF3B core in complex with sudemycin D6 (form I) | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
6DER
 
 | | Crystal structure of Candida albicans acetohydroxyacid synthase in complex with the herbicide metosulam | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Guddat, L.W. | | Deposit date: | 2018-05-12 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.126 Å) | | Cite: | Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DEN
 
 | |
3T77
 
 | | S25-2- A(2-4)KDO disaccharide complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S25-2 FAB (IGG1K) heavy chain, ... | | Authors: | Nguyen, H.P, Seto, N.O, Mackenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2011-07-29 | | Release date: | 2011-08-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
6EI1
 
 | | Crystal structure of the covalent complex between deubiquitinase ZUFSP (ZUP1) and Ubiquitin-PA | | Descriptor: | GLYCEROL, MALONATE ION, Polyubiquitin-B, ... | | Authors: | Pichlo, C, Baumann, U, Hofmann, K, Hermanns, T. | | Deposit date: | 2017-09-16 | | Release date: | 2018-03-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A family of unconventional deubiquitinases with modular chain specificity determinants.
Nat Commun, 9, 2018
|
|
7OM5
 
 | | Anti-EGFR nanobody EgB4 | | Descriptor: | GLYCEROL, Nanobody EgB4, ZINC ION | | Authors: | Zeronian, M.R, Janssen, B.J.C. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the non-inhibitory mechanism of the anti-EGFR EgB4 nanobody.
Bmc Mol Cell Biol, 23, 2022
|
|
7OM4
 
 | |
6E5T
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiorcin (Abn-CBDO) | | Descriptor: | (1'R,2'R)-5',6-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6EEC
 
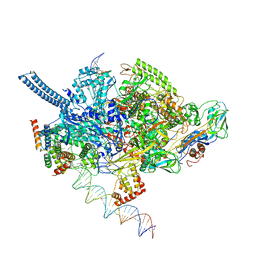 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter captured by Corallopyronin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
7OVO
 
 | | Heterodimeric murine tRNA-guanine transglycosylase in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Queuine tRNA-ribosyltransferase accessory subunit 2, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
6E5L
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-20 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6E5W
 
 | | Crystal structure of human cellular retinol binding protein 3 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, GLYCEROL, Retinol-binding protein 5 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
7OWZ
 
 | | Heterodimeric murine tRNA-guanine transglycosylase in complex with queuine and in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
6E6K
 
 | | Crystal structure of human cellular retinol-binding protein 4 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinoid-binding protein 7 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6EN4
 
 | | SF3b core in complex with a splicing modulator | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural Basis of Splicing Modulation by Antitumor Macrolide Compounds.
Mol. Cell, 70, 2018
|
|
3T65
 
 | | S25-2- A(2-8)KDO disaccharide complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S25-2 FAB (IGG1K) heavy chain, ... | | Authors: | Nguyen, H.P, Seto, N.O, Mackenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
6FDK
 
 | |
6FKJ
 
 | | Tubulin-TUB075 complex | | Descriptor: | (5~{S})-2-[(~{E})-~{N}-(2-ethoxyphenyl)-~{C}-methyl-carbonimidoyl]-3-oxidanyl-5-phenyl-cyclohex-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Steinmetz, M.O, Priego, E.-M. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | High-affinity ligands of the colchicine domain in tubulin based on a structure-guided design.
Sci Rep, 8, 2018
|
|
6DEO
 
 | |
6DEQ
 
 | | Crystal structure of Candida albicans acetohydroxyacid synthase in complex with the herbicide penoxsulam | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-(2,2-difluoroethoxy)-N-(5,8-dimethoxy[1,2,4]triazolo[1,5-c]pyrimidin-2-yl)-6-(trifluoromethyl)benzenesulfonamide, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Guddat, L.W. | | Deposit date: | 2018-05-12 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E6M
 
 | | Crystal structure of human cellular retinol-binding protein 1 in complex with cannabidiorcin (CBDO) | | Descriptor: | (1'R,2'R)-4,5'-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,6-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6GJB
 
 | | Erk2 signalling protein | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[4-[1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-2-oxidanylidene-pyridin-4-yl]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Quantitation of ERK1/2 inhibitor cellular target occupancies with a reversible slow off-rate probe.
Chem Sci, 9, 2018
|
|
