6G30
 
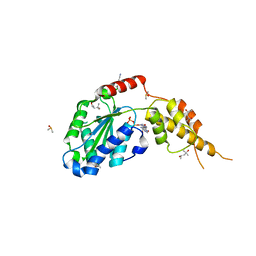 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.418 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G4F
 
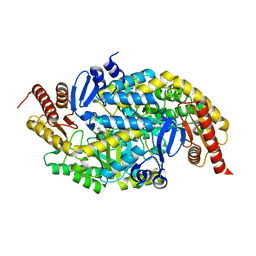 | |
6GDW
 
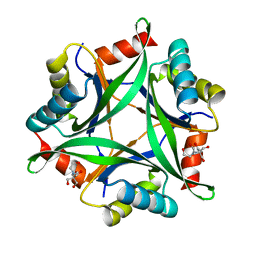 | |
5F3H
 
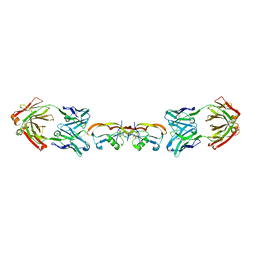 | | Structure of myostatin in complex with humanized RK35 antibody | | Descriptor: | Growth/differentiation factor 8, humanized RK35 antibody heavy chain, humanized RK35 antibody light chain | | Authors: | Parris, K.D, Mosyak, L. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody.
Mabs, 8, 2016
|
|
6GDV
 
 | | Structure of CutA from Synechococcus elongatus PCC7942 complexed with Bis-Tris molecule | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
6GDX
 
 | |
6PX2
 
 | | Acropora millepora GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Brandt, G.S, Fields, P.A. | | Deposit date: | 2019-07-24 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermal stability and structure of glyceraldehyde-3-phosphate dehydrogenase from the coral Acropora millepora.
Rsc Adv, 11, 2021
|
|
5F3B
 
 | | Structure of myostatin in complex with chimeric RK35 antibody | | Descriptor: | GLYCEROL, Growth/differentiation factor 8, RK35 Chimeric antibody heavy chain, ... | | Authors: | Parris, K.D, Mosyak, L. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody.
Mabs, 8, 2016
|
|
6G2W
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.678 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G2V
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G2Y
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G4B
 
 | |
6G4D
 
 | |
6G2Z
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G4E
 
 | |
5EUJ
 
 | | PYRUVATE DECARBOXYLASE FROM ZYMOBACTER PALMAE | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pyruvate decarboxylase from Zymobacter palmae.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6GDU
 
 | | Structure of CutA from Synechococcus elongatus PCC7942 | | Descriptor: | Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
6GIR
 
 | | Arabidopsis thaliana cytosolic seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase, cytoplasmic | | Authors: | Kekez, I, Kekez, M, Rokov-Plavec, J, Matkovic-Calogovic, D. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Arabidopsis seryl-tRNA synthetase: the first crystal structure and novel protein interactor of plant aminoacyl-tRNA synthetase.
FEBS J., 286, 2019
|
|
6GUJ
 
 | | Molybdenum storage protein with two occupied ATP binding sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MOLYBDATE ION, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
6GG1
 
 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
6Q3U
 
 | | Gly52Ala mutant of arginine-bound ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Balasco, N, Smaldone, G, Ruggiero, A, Vitagliano, L. | | Deposit date: | 2018-12-04 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The characterization of Thermotoga maritima Arginine Binding Protein variants demonstrates that minimal local strains have an important impact on protein stability.
Sci Rep, 9, 2019
|
|
1PS3
 
 | | Golgi alpha-mannosidase II in complex with kifunensine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase II, ... | | Authors: | Shah, N, Kuntz, D.A, Rose, D.R. | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of Kifunensine and 1-Deoxymannojirimycin Binding to Class I and II alpha-Mannosidases Demonstrates Different Saccharide Distortions in Inverting and Retaining Catalytic Mechanisms
Biochemistry, 42, 2003
|
|
6GX4
 
 | | The molybdenum storage protein: with ATP/Mn2+ and with POM clusters formed under in vitro conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, MOLYBDATE ION, ... | | Authors: | Poppe, J, Bruenle, S, Hail, R, Ermler, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
6GU5
 
 | | Mosto containing the core POM clusters | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MOLYBDATE ION, Molybdenum storage protein subunit alpha, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
6H2Z
 
 | | The crystal structure of human carbonic anhydrase II in complex with 4-(4-phenylpiperidine-1-carbonyl)benzenesulfonamide. | | Descriptor: | 4-(4-phenylpiperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
