3GZU
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2XJ8
 
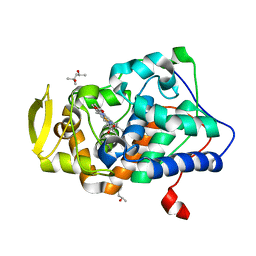 | | The structure of ferrous cytochrome c peroxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
1M1Q
 
 | | P222 oxidized structure of the tetraheme cytochrome c from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
6ZJD
 
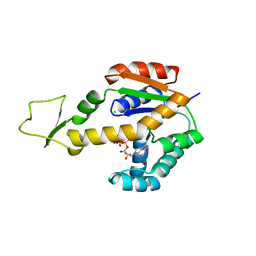 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
1ML7
 
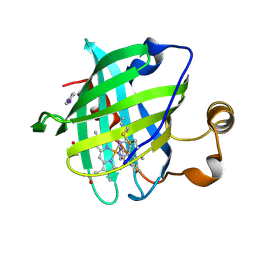 | | Crystal structure of nitrophorin 4 complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, 5,8-DIMETHYL-1,2,3,4-TETRAVINYLPORPHINE-6,7-DIPROPIONIC ACID FERROUS COMPLEX, nitrophorin 4 | | Authors: | Berry, R.E, Ding, X.D, Weichsel, A, Montfort, W.R, Walker, F.A. | | Deposit date: | 2002-08-30 | | Release date: | 2002-09-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Axial ligand complexes of the Rhodnius nitrophorins: reduction potentials, binding constants, EPR spectra, and structures of the 4-iodopyrazole and imidazole complexes of NP4
J.Biol.Inorg.Chem., 9, 2004
|
|
6ZLF
 
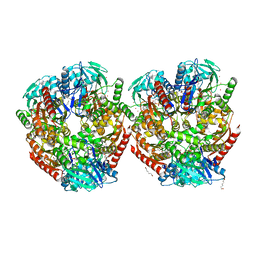 | | Aerobic crystal structure of F420H2-Oxidase from Methanothermococcus thermolithotrophicus at 1.8A resolution under 125 bars of krypton | | Descriptor: | CHLORIDE ION, Coenzyme F420H2 oxidase (FprA), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Engilberge, S, Wagner, T, Carpentier, P, Girard, E, Shima, S. | | Deposit date: | 2020-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Krypton-derivatization highlights O 2 -channeling in a four-electron reducing oxidase.
Chem.Commun.(Camb.), 56, 2020
|
|
2XIL
 
 | | The structure of cytochrome c peroxidase Compound I | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XI6
 
 | | The structure of ascorbate peroxidase Compound I | | Descriptor: | ASCORBATE PEROXIDASE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
1M7R
 
 | | Crystal Structure of Myotubularin-related Protein-2 (MTMR2) Complexed with Phosphate | | Descriptor: | Myotubularin-related Protein-2, PHOSPHATE ION | | Authors: | Begley, M.J, Taylor, G.S, Kim, S.-A, Veine, D.M, Dixon, J.E, Stuckey, J.A. | | Deposit date: | 2002-07-22 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome
Mol.Cell, 12, 2003
|
|
2XIH
 
 | | The structure of ascorbate peroxidase Compound III | | Descriptor: | ASCORBATE PEROXIDASE, OXYGEN MOLECULE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
6Z84
 
 | | CK2 alpha bound to chemical probe SGC-CK2-1 derivative | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[1-[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]indol-6-yl]ethanamide | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
3H8C
 
 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors (compound 14) | | Descriptor: | Cathepsin L1, N-(biphenyl-4-ylacetyl)-S-methyl-L-cysteinyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Combined Crystallographic and Molecular Dynamics Study of Cathepsin L Retrobinding Inhibitors
J.Med.Chem., 2009
|
|
6ZHO
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer with bound high affinity inhibitor | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Southall, S.M. | | Deposit date: | 2020-06-23 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Drug Discovery ofN-((R)-3-(7-Methyl-1H-indazol-5-yl)-1-oxo-1-(((S)-1-oxo-3-(piperidin-4-yl)-1-(4-(pyridin-4-yl)piperazin-1-yl)propan-2-yl)amino)propan-2-yl)-2'-oxo-1',2'-dihydrospiro[piperidine-4,4'-pyrido[2,3-d][1,3]oxazine]-1-carboxamide (HTL22562): A Calcitonin Gene-Related Peptide Receptor Antagonist for Acute Treatment of Migraine.
J.Med.Chem., 63, 2020
|
|
1M6E
 
 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
7LIR
 
 | | Structure of the invertebrate ALK GRD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALK tyrosine kinase receptor homolog scd-2, ... | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-01-27 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
6ZCV
 
 | |
7LTT
 
 | | SAMHD1(113-626) H206R D207N R366C | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Temple, J.T, Bowen, N.E. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization explains loss of dNTPase activity of the cancer-specific R366C/H mutant SAMHD1 proteins.
J.Biol.Chem., 297, 2021
|
|
6YSK
 
 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | Descriptor: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
2XJ5
 
 | | The structure of cytochrome c peroxidase Compound II | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
6ZHJ
 
 | | 3D electron diffraction structure of thermolysin from Bacillus thermoproteolyticus | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.26 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZK8
 
 | | Native crystal structure of anaerobic F420H2-Oxidase from Methanothermococcus thermolithotrophicus at 1.8A resolution | | Descriptor: | Coenzyme F420H2 oxidase (FprA), DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Engilberge, S, Wagner, T, Carpentier, P, Girard, E, Shima, S. | | Deposit date: | 2020-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Krypton-derivatization highlights O 2 -channeling in a four-electron reducing oxidase.
Chem.Commun.(Camb.), 56, 2020
|
|
6DPU
 
 | | Undecorated GMPCPP microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Separating the effects of nucleotide and EB binding on microtubule structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6Z43
 
 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
1M9N
 
 | | CRYSTAL STRUCTURE OF THE HOMODIMERIC BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME AVIAN ATIC IN COMPLEX WITH AICAR AND XMP AT 1.93 ANGSTROMS. | | Descriptor: | AICAR TRANSFORMYLASE-IMP CYCLOHYDROLASE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Wolan, D.W, Greasly, S.E, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2002-07-29 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Avian AICAR Transformylase Mechanism.
Biochemistry, 41, 2002
|
|
6Z2O
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, O-glycan protease, ZINC ION | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
