1B4N
 
 | | FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE FROM PYROCOCCUS FURIOSUS, COMPLEXED WITH GLUTARATE | | Descriptor: | CALCIUM ION, FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE, GLUTARIC ACID, ... | | Authors: | Hu, Y.L, Faham, S, Roy, R, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1998-12-24 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: the 1.85 A resolution crystal structure and its mechanistic implications.
J.Mol.Biol., 286, 1999
|
|
6LUQ
 
 | | Haloperidol bound D2 dopamine receptor structure inspired discovery of subtype selective ligands | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, OLEIC ACID, chimera of D(2) dopamine receptor and Endolysin | | Authors: | Fan, L, Tan, L, Chen, Z, Qi, J, Nie, F, Luo, Z, Cheng, J, Wang, S. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Haloperidol bound D2dopamine receptor structure inspired the discovery of subtype selective ligands.
Nat Commun, 11, 2020
|
|
6I1H
 
 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein,Penicillin-binding protein | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem
To Be Published
|
|
2BRN
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | (2R)-1-[(5,6-DIPHENYL-7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)AMINO]PROPAN-2-OL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
5W6K
 
 | | Structure of mutant Taq Polymerase incorporating unnatural base pairs Z:P | | Descriptor: | (1R)-1-[6-amino-5-(dihydroxyamino)-2-hydroxypyridin-3-yl]-1,4-anhydro-2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-erythro-pentitol, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA (5'-D(P*(1WA)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Singh, I, Georgiadis, M.M. | | Deposit date: | 2017-06-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Snapshots of an evolved DNA polymerase pre- and post-incorporation of an unnatural nucleotide.
Nucleic Acids Res., 46, 2018
|
|
4RQK
 
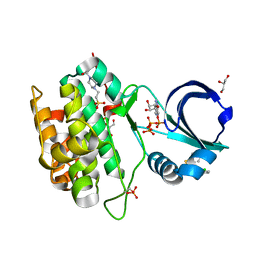 | |
7A3H
 
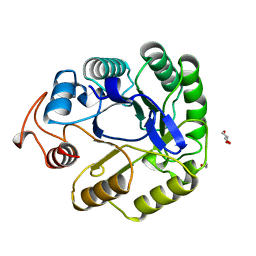 | | NATIVE ENDOGLUCANASE CEL5A CATALYTIC CORE DOMAIN AT 0.95 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, ETHANOL, GLYCEROL | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-08-05 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
2C2N
 
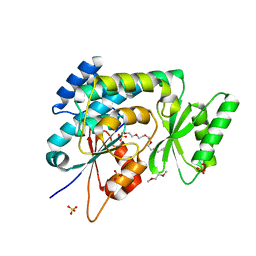 | | Structure of human mitochondrial malonyltransferase | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-(2-ETHOXYETHOXY)ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ... | | Authors: | Wu, X, Bunkoczi, G, Smee, C, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2005-09-29 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Different Specificities of Acyltransferases Associated with the Human Cytosolic and Mitochondrial Fatty Acid Synthases.
Chem.Biol., 16, 2009
|
|
6GCO
 
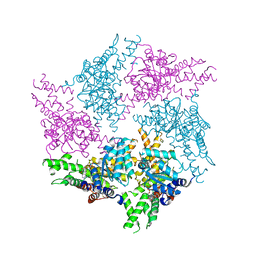 | | Truncated FtsH from A. aeolicus in P312 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, ZINC ION | | Authors: | Uthoff, M, Baumann, U. | | Deposit date: | 2018-04-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Conformational flexibility of pore loop-1 gives insights into substrate translocation by the AAA+protease FtsH.
J. Struct. Biol., 204, 2018
|
|
4RQV
 
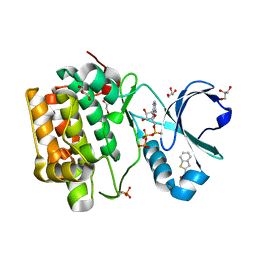 | |
6IUP
 
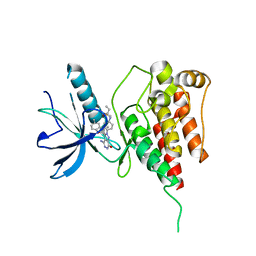 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6GED
 
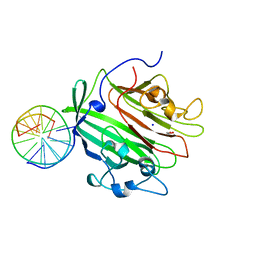 | | Adhesin domain of PrgB from Enterococcus faecalis bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*GP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*CP*GP*GP*CP*CP*CP*G)-3'), ... | | Authors: | Schmitt, A, Berntsson, R.P.A. | | Deposit date: | 2018-04-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction.
Mol. Microbiol., 109, 2018
|
|
9J4Y
 
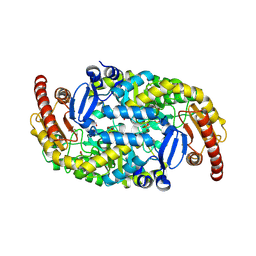 | | Crystal Structure of the L322F mutant of Omega Transaminase TA_2799 from Pseudomonas putida KT2440 | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase, class III, ... | | Authors: | Das, P, Bhaumik, P. | | Deposit date: | 2024-08-10 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights and rational design of Pseudomonasputida KT2440 omega transaminases for enhanced biotransformation of (R)-PAC to (1R, 2S)-Norephedrine.
J.Biol.Chem., 301, 2025
|
|
1CBQ
 
 | | CRYSTAL STRUCTURE OF CELLULAR RETINOIC-ACID-BINDING PROTEINS I AND II IN COMPLEX WITH ALL-TRANS-RETINOIC ACID AND A SYNTHETIC RETINOID | | Descriptor: | 6-(2,3,4,5,6,7-HEXAHYDRO-2,4,4-TRIMETHYL-1-METYLENEINDEN-2-YL)-3-METHYLHEXA-2,4-DIENOIC ACID, CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II, PHOSPHATE ION | | Authors: | Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1994-09-28 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of cellular retinoic acid binding proteins I and II in complex with all-trans-retinoic acid and a synthetic retinoid.
Structure, 2, 1994
|
|
1BRY
 
 | | BRYODIN TYPE I RIP | | Descriptor: | BRYODIN I | | Authors: | Klei, H.E, Chang, C.Y. | | Deposit date: | 1997-02-14 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular, biological, and preliminary structural analysis of recombinant bryodin 1, a ribosome-inactivating protein from the plant Bryonia dioica.
Biochemistry, 36, 1997
|
|
7VLU
 
 | | Structure of SUR2A in complex with Mg-ATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
2BRO
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | (2R)-3-{[(4Z)-5,6-DIPHENYL-6,7-DIHYDRO-4H-PYRROLO[2,3-D]PYRIMIDIN-4-YLIDENE]AMINO}PROPANE-1,2-DIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
4RUR
 
 | | Yeast 20S proteasome in complex with the alkaloid indolo-phakellin (4) | | Descriptor: | (2E,3aR,14aS)-9-bromo-2-imino-1,2,3,5,6,14a-hexahydro-4H,8H-imidazo[4',5':5,6]pyrrolo[1',2':4,5]pyrazino[1,2-a]indol-8-one, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Beck, P, Lansdell, T.A, Hewlett, N.M, Tepe, J.J, Groll, M. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Indolo-Phakellins as beta 5-Specific Noncovalent Proteasome Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5ZL1
 
 | | Hexameric structure of copper-containing nitrite reductase of an anammox organism KSU-1 | | Descriptor: | CALCIUM ION, COPPER (II) ION, Putative copper-type nitrite reductase, ... | | Authors: | Hira, D, Matsumura, M, Kitamura, R, Fujii, T. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hexameric structure of copper-containing nitrite reductase of an anammox organism KSU-1
To Be Published
|
|
6TTM
 
 | | Hyoscyamine 6-hydroxylase in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyoscyamine 6 beta-hydroxylase, ... | | Authors: | Kluza, A, Kurpiewska, K, Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
7B6J
 
 | | Crystal structure of MurE from E.coli in complex with minifrag succinimide | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
1BWF
 
 | | ESCHERICHIA COLI GLYCEROL KINASE MUTANT WITH BOUND ATP ANALOG SHOWING SUBSTANTIAL DOMAIN MOTION | | Descriptor: | GLYCEROL, GLYCEROL KINASE, MAGNESIUM ION, ... | | Authors: | Bystrom, C.E, Pettigrew, D.W, Branchaud, B.P, Remington, S.J. | | Deposit date: | 1998-09-23 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Escherichia coli glycerol kinase variant S58-->W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion.
Biochemistry, 38, 1999
|
|
8TI4
 
 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | GLYCEROL, IgG1 Fab heavy chain, mutated to promote correct pairing, ... | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
4YD3
 
 | | Endothiapepsin in complex with fragment 224 | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.248 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6N91
 
 | | Crystal Structure of Adenosine Deaminase from Vibrio cholerae Complexed with Pentostatin (Deoxycoformycin) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCOFORMYCIN, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Welk, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Adenosine Deaminase from Vibrio cholerae Complexed with Pentostatin (Deoxycoformycin) (CASP target)
To Be Published
|
|
