2VKP
 
 | | Crystal structure of BTB domain from BTBD6 | | Descriptor: | 1,2-ETHANEDIOL, BTB/POZ DOMAIN-CONTAINING PROTEIN 6, POTASSIUM ION | | Authors: | Cooper, C.D.O, Pike, A.C.W, Salah, E, Filippakopoulos, P, Bunkoczi, G, Elkins, J.M, von Delft, F, Gileadi, O, Edwards, A, Weigelt, J, Arrowsmith, C.H, Knapp, S. | | Deposit date: | 2007-12-21 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Btb Domain from Btbd6
To be Published
|
|
4FNY
 
 | | Crystal structure of the R1275Q anaplastic lymphoma kinase catalytic domain in complex with a benzoxazole inhibitor | | Descriptor: | ALK tyrosine kinase receptor, N-(4-chlorophenyl)-5-[(6,7-dimethoxyquinolin-4-yl)oxy]-1,3-benzoxazol-2-amine | | Authors: | Whittington, D.A, Epstein, L.F, Chen, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The R1275Q Neuroblastoma Mutant and Certain ATP-competitive Inhibitors Stabilize Alternative Activation Loop Conformations of Anaplastic Lymphoma Kinase.
J.Biol.Chem., 287, 2012
|
|
1MU0
 
 | | Crystal Structure of the Tricorn Interacting Factor F1 Complex with PCK | | Descriptor: | (2R,3S)-3-AMINO-1-CHLORO-4-PHENYL-BUTAN-2-OL, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
5JH7
 
 | | Tubulin-Eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Doodhi, H, Prota, A.E, Rodriguez-Garcia, R, Xiao, H, Custar, D.W, Bargsten, K, Katrukha, E.A, Hilbert, M, Hua, S, Jiang, K, Grigoriev, I, Yang, C.-P.H, Cox, D, Band Horwitz, S, Kapitein, L.C, Akhmanova, A, Steinmetz, M.O. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Termination of Protofilament Elongation by Eribulin Induces Lattice Defects that Promote Microtubule Catastrophes.
Curr.Biol., 26, 2016
|
|
6JAT
 
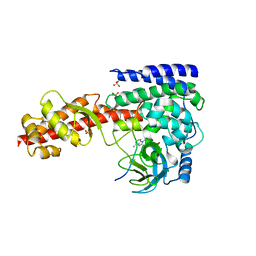 | | Crystal structure of SETD3 bound to Actin peptide and SFG | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Actin, gamma-enteric smooth muscle, ... | | Authors: | Li, H, Zheng, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Crystal structure of SETD3 bound to Actin peptide and SFG
to be published
|
|
3L2K
 
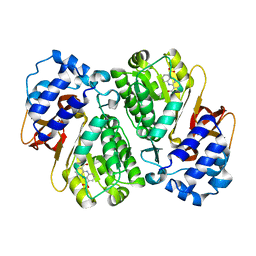 | | Structure of phenazine antibiotic biosynthesis protein with substrate | | Descriptor: | EhpF, phenazine-1,6-dicarboxylic acid | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-12-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the D-alanylgriseoluteic acid biosynthetic protein EhpF, an atypical member of the ANL superfamily of adenylating enzymes.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4Y4Z
 
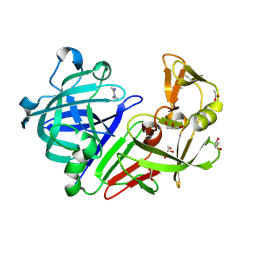 | | Endothiapepsin in complex with fragment 73 | | Descriptor: | (1Z)-N,N-diethylethanimidamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y5K
 
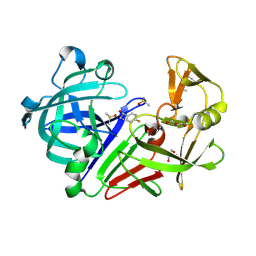 | | Endothiapepsin in complex with fragment 274 | | Descriptor: | (1S)-2-amino-1-(4-fluorophenyl)ethanol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
to be published
|
|
9BXH
 
 | |
9BXK
 
 | | OvsM from Marinimicrobium koreense, an ovoselenol-biosynthetic N-methyltransferase in complex with 5-selenohistidine and SAH | | Descriptor: | 1,2-ETHANEDIOL, 5-selanyl-L-histidine, 5-selenohistidine N-methyltransferase OvsM, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-05-22 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and functional analysis of SAM-dependent N-methyltransferases involved in ovoselenol and ovothiol biosynthesis.
Structure, 33, 2025
|
|
2VCX
 
 | | Complex structure of prostaglandin D2 synthase at 2.1A. | | Descriptor: | GLUTATHIONE, GLUTATHIONE-REQUIRING PROSTAGLANDIN D SYNTHASE, MAGNESIUM ION, ... | | Authors: | Hohwy, M, Spadola, L, Lundquist, B, von Wachenfeldt, K, Persdotter, S, Hawtin, P, Dahmen, J, Groth-Clausen, I, Folmer, R.H.A, Edman, K. | | Deposit date: | 2007-09-27 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel Prostaglandin D Synthase Inhibitors Generated by Fragment-Based Drug Design.
J.Med.Chem., 51, 2008
|
|
9BXJ
 
 | |
4MZS
 
 | |
8VTW
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-128 and protein Y at 2.35A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-cyclopropyl-7-(4-{4-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]butyl}piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
2UZF
 
 | | Crystal structure of Staphylococcus aureus 1,4-dihydroxy-2-naphthoyl CoA synthase (MenB) in complex with acetoacetyl CoA | | Descriptor: | ACETOACETYL-COENZYME A, NAPHTHOATE SYNTHASE | | Authors: | Ulaganathan, V, Buetow, L, Hunter, W.N. | | Deposit date: | 2007-04-27 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Staphylococcus Aureus1,4-Dihydroxy-2-Naphthoyl-Coa Synthase (Menb) in Complex with Acetoacetyl-Coa.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4RWH
 
 | | Crystal structure of T cell costimulatory ligand B7-1 (CD80) | | Descriptor: | T-lymphocyte activation antigen CD80 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2014-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of T cell costimulatory ligand B7-1 (CD80)
To be Published
|
|
5F9S
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.7 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
4RXG
 
 | | Fructose-6-phosphate aldolase Q59E from E.coli | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL, PENTAETHYLENE GLYCOL | | Authors: | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
1N1Z
 
 | | (+)-Bornyl Diphosphate Synthase: Complex with Mg and pyrophosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6TNZ
 
 | | Human polymerase delta-FEN1-PCNA toolbelt | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-12-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6EKN
 
 | | Crystal structure of MMP12 in complex with inhibitor BE7. | | Descriptor: | (2~{S})-2-[2-[4-(4-methoxyphenyl)phenyl]sulfonylphenyl]pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Ciccone, L, Tepshi, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
6THW
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 7-fluoranyl-4-(1-methylcyclopropyl)oxy-~{N}-[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]-6-(2-methylpyrimidin-5-yl)pyrido[3,2-d]pyrimidin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
9BUX
 
 | | Structure of GGCX-BGP-menaquinone-4 epoxide complex | | Descriptor: | (1aR,7aS)-1a-methyl-7a-[(2E,6E,10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl]-1a,7a-dihydronaphtho[2,3-b]oxirene-2,7-dione, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Qi, X. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure and mechanism of vitamin-K-dependent gamma-glutamyl carboxylase.
Nature, 639, 2025
|
|
7LHZ
 
 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid (compound 25) | | Descriptor: | (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*AP*TP*CP*AP*TP*AP*CP*AP*AP*CP*GP*TP*AP*A)-3'), ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2021-01-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
5B7V
 
 | | Human FGFR1 kinase in complex with CH5183284 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, [5-amino-1-(2-methyl-1H-benzimidazol-6-yl)-1H-pyrazol-4-yl](1H-indol-2-yl)methanone | | Authors: | Fukami, T.A, Lukacs, C.M, Janson, C. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor
Mol.Cancer Ther., 13, 2014
|
|
