6EPZ
 
 | |
9D9N
 
 | | Crystal structure of NDM-1 complexed with compound 5 | | Descriptor: | GLYCEROL, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2024-08-21 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Hit to Lead Optimization of Heteroaryl Phosphonates Reversible, Broad-Spectrum Inhibitors of Serine and Metallo Carbapenemases.
To Be Published
|
|
2VEO
 
 | | X-ray structure of Candida antarctica lipase A in its closed state. | | Descriptor: | GLYCEROL, LIPASE A, TETRAETHYLENE GLYCOL, ... | | Authors: | Ericsson, D.J, Kasrayan, A, Johansson, P, Bergfors, T, Sandstrom, A.G, Backvall, J.E, Mowbray, S.L. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure of Candida Antarctica Lipase a Shows a Novel Lid Structure and a Likely Mode of Interfacial Activation.
J.Mol.Biol., 376, 2008
|
|
2VCX
 
 | | Complex structure of prostaglandin D2 synthase at 2.1A. | | Descriptor: | GLUTATHIONE, GLUTATHIONE-REQUIRING PROSTAGLANDIN D SYNTHASE, MAGNESIUM ION, ... | | Authors: | Hohwy, M, Spadola, L, Lundquist, B, von Wachenfeldt, K, Persdotter, S, Hawtin, P, Dahmen, J, Groth-Clausen, I, Folmer, R.H.A, Edman, K. | | Deposit date: | 2007-09-27 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel Prostaglandin D Synthase Inhibitors Generated by Fragment-Based Drug Design.
J.Med.Chem., 51, 2008
|
|
9DAG
 
 | | Crystal structure of NDM-1 complexed with compound 13 | | Descriptor: | GLYCEROL, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2024-08-22 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hit to Lead Optimization of Heteroaryl Phosphonates Reversible, Broad-Spectrum Inhibitors of Serine and Metallo Carbapenemases.
To Be Published
|
|
1RTU
 
 | | USTILAGO SPHAEROGENA RIBONUCLEASE U2 | | Descriptor: | RIBONUCLEASE U2, SULFATE ION | | Authors: | Noguchi, S, Satow, Y, Uchida, T, Sasaki, C, Matsuzaki, T. | | Deposit date: | 1995-05-12 | | Release date: | 1996-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ustilago sphaerogena ribonuclease U2 at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
8UG2
 
 | | Crystal structure of de novo designed metal-controlled heterodimer of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G MCHeT_A + MCHeT_C | | Descriptor: | 1,2-ETHANEDIOL, Beta 1 domain of streptococcal protein G (G beta 1) MCHetA, Beta 1 domain of streptococcal protein G (G beta 1) MCHetC, ... | | Authors: | Mealka, M, Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Design of Protein Heterodimers Utilizing Metal-Ligand and
Salt-Bridge Interactions
To Be Published
|
|
9DE3
 
 | | Crystal structure of NDM-1 complexed with compound 28 | | Descriptor: | Metallo-beta-lactamase type 2, ZINC ION, {[1-(cyclopentylmethyl)-3-methyl-2-oxo-7-(pyrrolidin-1-yl)-1,2-dihydroquinolin-4-yl]methyl}phosphonic acid | | Authors: | Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2024-08-28 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Hit to Lead Optimization of Heteroaryl Phosphonates Reversible, Broad-Spectrum Inhibitors of Serine and Metallo Carbapenemases.
To Be Published
|
|
5L6I
 
 | | Uba1 in complex with Ub-MLN4924 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
8BCA
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 26 | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-~{N}-methyl-4-(methylamino)benzenesulfonamide, GLYCEROL, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7RYN
 
 | | CD1a-sulfatide-gdTCR complex | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
4O8J
 
 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii, in complex with rACAAA3'phosphate and adenine. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE, RNA, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
9LYG
 
 | | Crystal structure of FKBP12 complexed with Small Molecule Anchor for Protein-201 | | Descriptor: | 5-[(2~{S})-1-cyclohexylsulfonylpiperidin-2-yl]-3-[3-(3,4-dimethoxyphenyl)propyl]-1,2,4-oxadiazole, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Kato, S, Tsuchikawa, H, Katoh, A, Matsuoka, S, Sugiyama, S. | | Deposit date: | 2025-02-20 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of FKBP12 complexed with Small Molecule Anchor for Protein-201
To Be Published
|
|
7OF3
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
3K9T
 
 | |
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
5G3J
 
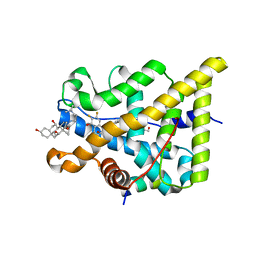 | | Discovery of New Selective Glucocorticoid Receptor Agonist Leads | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5-[[(1S,2R,4R)-4-ethyl-6,7-bis(fluoranyl)-2,5-bis(oxidanyl)-2-(trifluoromethyl)-3,4-dihydro-1H-naphthalen-1-yl]amino]-1H-quinolin-2-one, ... | | Authors: | Berger, M, Edman, K, Wissler, L, Neuhaus, R, Rehwinkel, H, Schacke, H, Jaroch, S. | | Deposit date: | 2016-04-27 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of New Selective Glucocorticoid Receptor Agonist Leads.
Bioorg.Med.Chem.Lett., 27, 2017
|
|
6CAX
 
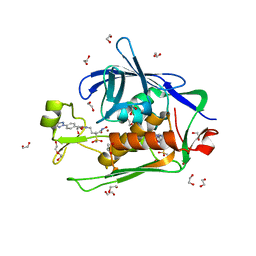 | |
5D3N
 
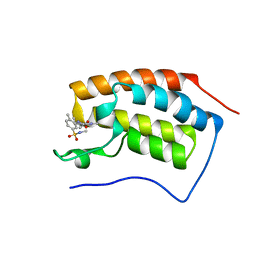 | | First bromodomain of BRD4 bound to inhibitor XD40 | | Descriptor: | 4-acetyl-3-ethyl-5-methyl-N-[2-methyl-5-(methylsulfamoyl)phenyl]-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
6LE7
 
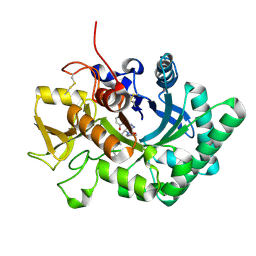 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2 | | Descriptor: | (4R)-3-(2-hydroxyphenyl)-4-(3-methoxy-4-propoxy-phenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, Probable endochitinase | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85753441 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2
To Be Published
|
|
5DB3
 
 | | Menin in complex with MI-574 | | Descriptor: | 6-methoxy-4-methyl-1-(1H-pyrazol-4-ylmethyl)-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
6C44
 
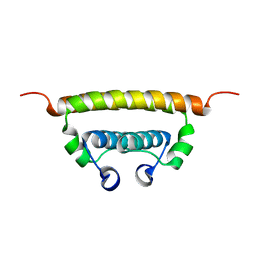 | | Zika virus capsid protein | | Descriptor: | Capsid protein | | Authors: | Morando, M.A, Barbosa, G.M, Cruz-Oliveira, C, Da Poian, A.T, Almeida, F.C.L. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamics of Zika Virus Capsid Protein in Solution: The Properties and Exposure of the Hydrophobic Cleft Are Controlled by the alpha-Helix 1 Sequence.
Biochemistry, 58, 2019
|
|
1RBV
 
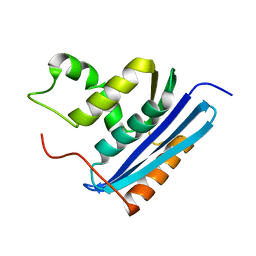 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
3DTF
 
 | |
