4ZEY
 
 | |
4PTV
 
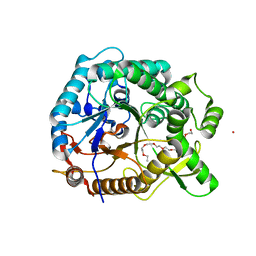 | | Halothermothrix orenii beta-glucosidase A, thiocellobiose complex | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CESIUM ION, Glycoside hydrolase family 1, ... | | Authors: | Hassan, N, Nguyen, T.H, Kori, L.D, Patel, B.K.C, Haltrich, D, Divne, C, Tan, T.C. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of a thermostable beta-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4Z86
 
 | |
1AFE
 
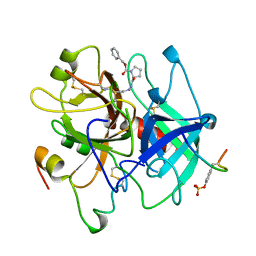 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
4PC7
 
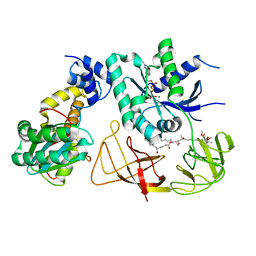 | | Elongation factor Tu:Ts complex in a near GTP conformation. | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, Elongation factor Ts, Elongation factor Tu 1, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.6003 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
5TRY
 
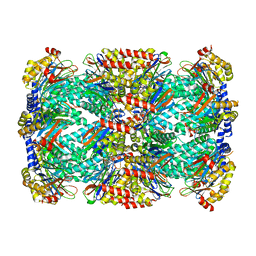 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2206 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-3-methoxy-1-(naphthalen-1-ylmethylamino)-1-oxidanylidene-propan-2-yl]-4-oxidanylidene-2-(3-phenylpropanoylamino)-4-piperidin-1-yl-butanamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.000008 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
155L
 
 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
164L
 
 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
5WHC
 
 | | USP7 in complex with Cpd2 (4-(3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl)phenol) | | Descriptor: | 4-[3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl]phenol, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-07-16 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of Ubiquitin Specific Protease 7 (USP7) Using Integrated NMR and in Silico Techniques.
J. Med. Chem., 60, 2017
|
|
6UOK
 
 | |
6IUO
 
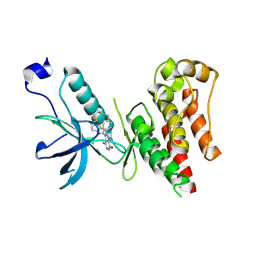 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
5WZO
 
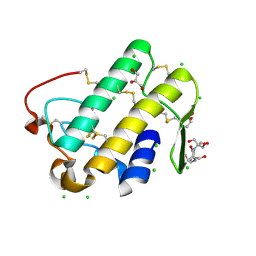 | | Crystal structure of human secreted phospholipase A2 group IIE, crystallized with calcium | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5A2S
 
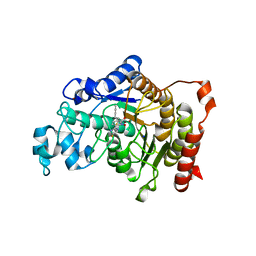 | | Potent, selective and CNS-penetrant tetrasubstituted cyclopropane class IIa histone deacetylase (HDAC) inhibitors | | Descriptor: | (1S,2S,3S)-1-fluoranyl-2-[4-(5-fluoranylpyrimidin-2-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, SODIUM ION, ... | | Authors: | Luckhurst, C.A, Breccia, P, Stott, A.J, Aziz, O, Birch, H, Burli, R.W, Hughes, S, Jarvis, R.E, Lamers, M, Leonard, P, Matthews, K.L, McAllister, G, Pollack, S, Saville-Stones, E, Wishart, G, Yates, D, Dominguez, C. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Potent, Selective, and Cns-Penetrant Tetrasubstituted Cyclopropane Class Iia Histone Deacetylase (Hdac) Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5AJA
 
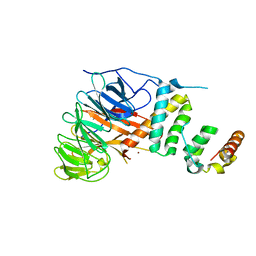 | | Crystal structure of mandrill SAMHD1 (amino acid residues 1-114) bound to Vpx isolated from mandrill and human DCAF1 (amino acid residues 1058-1396) | | Descriptor: | PROTEIN VPRBP, SAM DOMAIN AND HD DOMAIN-CONTAINING PROTEIN, VPX PROTEIN, ... | | Authors: | Schwefel, D, Boucherit, V.C, Christodoulou, E, Walker, P.A, Stoye, J.P, Bishop, K.N, Taylor, I.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Molecular Determinants for Recognition of Divergent Samhd1 Proteins by the Lentiviral Accessory Protein Vpx.
Cell Host Microbe., 17, 2015
|
|
4P8W
 
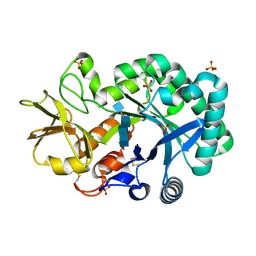 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc4) were solved to resolutions of 1.9 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
163L
 
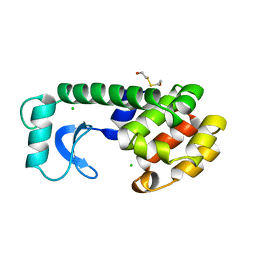 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
4CIA
 
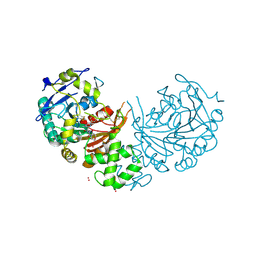 | | Crystal structure of cathepsin A, complexed with compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, LYSOSOMAL PROTECTIVE PROTEIN, ... | | Authors: | Schreuder, H.A, Liesum, A, Kroll, K, Boehnisch, B, Buning, C, Ruf, S, Buning, C, Sadowski, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of cathepsin A, a novel target for the treatment of cardiovascular diseases.
Biochem. Biophys. Res. Commun., 445, 2014
|
|
3JZJ
 
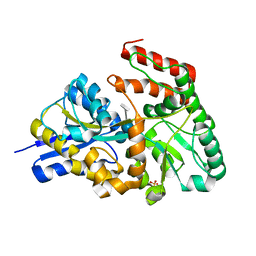 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose/maltose binding protein GacH, SULFATE ION | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
4ZFZ
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with myristoylated 5-mer lipopeptide derived from SIV Nef protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-mer lipopeptide from Protein Nef, ... | | Authors: | Morita, D, Sugita, M. | | Deposit date: | 2015-04-22 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of the N-myristoylated lipopeptide-bound MHC class I complex
Nat Commun, 7, 2016
|
|
5AJX
 
 | | Human PFKFB3 in complex with an indole inhibitor 3 | | Descriptor: | (2S)-N-[4-[3-cyano-1-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]indol-5-yl]oxyphenyl]pyrrolidine-2-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
1A2U
 
 | | STAPHYLOCOCCAL NUCLEASE, V23C VARIANT, COMPLEX WITH 1-N-BUTANE THIOL AND 3',5'-THYMIDINE DIPHOSPHATE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-11 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
4Z0I
 
 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
1A3Q
 
 | | HUMAN NF-KAPPA-B P52 BOUND TO DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*CP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B P52) | | Authors: | Cramer, P, Larson, C.J, Verdine, G.L, Muller, C.W. | | Deposit date: | 1998-01-23 | | Release date: | 1998-06-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution.
EMBO J., 16, 1997
|
|
5FPL
 
 | | Crystal structure of human JARID1B in complex with CCT363901 | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-(2-azanylethyl)pyrazol-1-yl]-3H-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Yann-Vai, L.B, Nowak, R, Johansson, C, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Huber, K, Oppermann, U. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 8-Substituted Pyrido[3,4-D]Pyrimidin-4(3H)-One Derivatives as Potent, Cell Permeable, Kdm4 (Jmjd2) and Kdm5 (Jarid1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
