1GKN
 
 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-16 | | Release date: | 2002-04-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
1YDI
 
 | |
1SS7
 
 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-23 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1DOV
 
 | |
1DA3
 
 | | THE CRYSTAL STRUCTURE OF THE TRIGONAL DECAMER C-G-A-T-C-G-6MEA-T-C-G: A B-DNA HELIX WITH 10.6 BASE-PAIRS PER TURN | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*AP*TP*CP*GP*(6MA)P*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Baikalov, I, Grzeskowiak, K, Yanagi, K, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-11-09 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn.
J.Mol.Biol., 231, 1993
|
|
2DFK
 
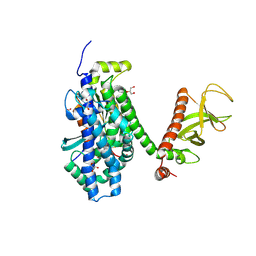 | | Crystal structure of the CDC42-Collybistin II complex | | Descriptor: | GLYCEROL, SULFATE ION, cell division cycle 42 isoform 1, ... | | Authors: | Xiang, S, Kim, E.Y, Connelly, J.J, Nassar, N, Kirsch, J, Winking, J, Schwarz, G, Schindelin, H. | | Deposit date: | 2006-03-02 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Cdc42 in Complex with Collybistin II, a Gephyrin-interacting Guanine Nucleotide Exchange Factor.
J.Mol.Biol., 359, 2006
|
|
1DK1
 
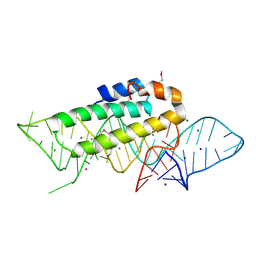 | | DETAILED VIEW OF A KEY ELEMENT OF THE RIBOSOME ASSEMBLY: CRYSTAL STRUCTURE OF THE S15-RRNA COMPLEX | | Descriptor: | 30S RIBOSOMAL PROTEIN S15, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Nikulin, A, Serganov, A, Ennifar, E, Tischenko, S, Nevskaya, N. | | Deposit date: | 1999-12-06 | | Release date: | 2000-04-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S15-rRNA complex.
Nat.Struct.Biol., 7, 2000
|
|
8A8J
 
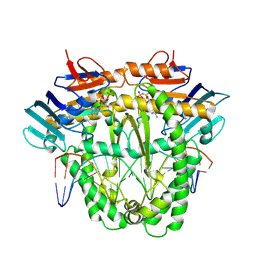 | | Complex of RecF and DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AB0
 
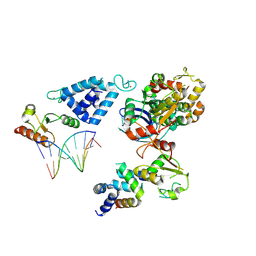 | | Complex of RecO-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA repair protein RecO, Oligo1, Oligo2, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A93
 
 | | Complex of RecF-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4LX3
 
 | | Conserved Residues that Modulate Protein trans-Splicing of Npu DnaE Split Intein | | Descriptor: | DNA polymerase III, alpha subunit, Nucleic acid binding, ... | | Authors: | Wu, Q, Gao, Z, Wei, Y, Ma, G, Zheng, Y, Dong, Y, Liu, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved residues that modulate protein trans-splicing of Npu DnaE split intein.
Biochem.J., 461, 2014
|
|
6TDM
 
 | | Bam_5920cDD 5919nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6ULG
 
 | | Cryo-EM structure of the FLCN-FNIP2-Rag-Ragulator complex | | Descriptor: | Folliculin, Folliculin-interacting protein 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shen, K, Rogala, K.B, Yu, Z.H, Sabatini, D.M. | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM Structure of the Human FLCN-FNIP2-Rag-Ragulator Complex.
Cell, 179, 2019
|
|
6TDD
 
 | | Bam_5924 docking domain | | Descriptor: | Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TDN
 
 | | Bam_5925cDD 5924nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6U3E
 
 | |
8AJA
 
 | | Structure of the Ancestral Scaffold Antigen-5 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
8AJL
 
 | | Structure of the Ancestral Scaffold Antigen-6 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
6U62
 
 | | Raptor-Rag-Ragulator complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rogala, K.B, Sabatini, D.M. | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for the docking of mTORC1 on the lysosomal surface.
Science, 366, 2019
|
|
6U6T
 
 | | Neuronal growth regulator 1 (NEGR1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuronal growth regulator 1, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
6U9V
 
 | |
6UJA
 
 | | Integrin alpha-v beta-8 in complex with pro-TGF-beta1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
6V6D
 
 | | Cryo-EM structure of human pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Deng, Z, He, Z, Yuan, P. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6U9W
 
 | |
3EGH
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1), the PP1 binding and PDZ domains of Spinophilin and the small natural molecular toxin Nodularin-R | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
