7L12
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 14 | | Descriptor: | (5S)-5-{3-[3-(benzyloxy)-5-chlorophenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L14
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 26 | | Descriptor: | 2-{3-[3-chloro-5-(cyclopropylmethoxy)phenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L13
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5S)-5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]phenyl}-2-oxo[2H-[1,3'-bipyridine]]-5-yl)pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L10
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 (2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 2-[3-(3,5-dichlorophenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-13 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
6SPH
 
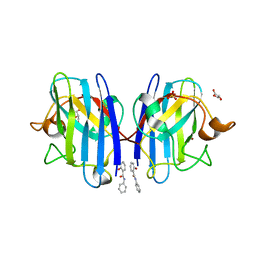 | | A4V MUTANT OF HUMAN SUPEROXIDE DISMUTASE 1 WITH EBSELEN BOND IN C2 SPACE GROUP | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-phenyl-2-selanylbenzamide, ... | | Authors: | Shahid, M, Chantadul, V, Amporndanai, K, Wright, G, Antonyuk, S, Hasnain, S. | | Deposit date: | 2019-09-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy.
Commun Biol, 3, 2020
|
|
7L11
 
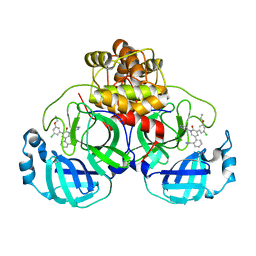 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 2-[3-(3-chloro-5-propoxyphenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
6T4B
 
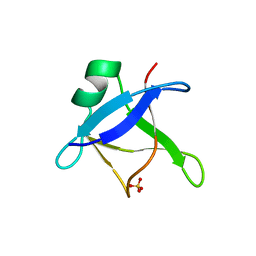 | | CRYSTAL STRUCTURE OF HUMAN TDP-43 N-TERMINAL DOMAIN AT 2.55 A RESOLUTION | | Descriptor: | SULFATE ION, TAR DNA-binding protein 43 | | Authors: | Watanabe, T.F, Wright, G.S.A, Amporndanai, K, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-10-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Purification and Structural Characterization of Aggregation-Prone Human TDP-43 Involved in Neurodegenerative Diseases.
Iscience, 23, 2020
|
|
6TCT
 
 | |
2LVG
 
 | |
3FVQ
 
 | | Crystal structure of the nucleotide binding domain FbpC complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Fe(3+) ions import ATP-binding protein fbpC | | Authors: | Newstead, S, Bilton, P, Carpenter, E.P, Campopiano, D, Iwata, S. | | Deposit date: | 2009-01-16 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into how nucleotide-binding domains power ABC transport.
Structure, 17, 2009
|
|
6HQ2
 
 | | Structure of EAL Enzyme Bd1971 - apo form | | Descriptor: | EAL Enzyme Bd1971 | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
3FIB
 
 | | RECOMBINANT HUMAN GAMMA-FIBRINOGEN CARBOXYL TERMINAL FRAGMENT (RESIDUES 143-411) BOUND TO CALCIUM AT PH 6.0: A FURTHER REFINEMENT OF PDB ENTRY 1FIB, AND DIFFERS FROM 1FIB BY THE MODELLING OF A CIS PEPTIDE BOND BETWEEN RESIDUES K338 AND C339 | | Descriptor: | CALCIUM ION, FIBRINOGEN GAMMA CHAIN RESIDUES | | Authors: | Pratt, K.P, Cote, H.C.F, Chung, D.W, Stenkamp, R.E, Davie, E.W. | | Deposit date: | 1997-07-14 | | Release date: | 1997-09-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2JLS
 
 | | Dengue virus 4 NS3 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLU
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', GLYCEROL, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLX
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA and ADP-Vanadate | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
8OF7
 
 | | Cyc15 Diels Alderase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Back, C.R, Barringer, R.W.L, Zorn, K, Manzo-Ruiz, M, Race, P.R. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Interrogation of an Enzyme Library Reveals the Catalytic Plasticity of Naturally Evolved [4+2] Cyclases.
Chembiochem, 24, 2023
|
|
8OE1
 
 | |
8OE5
 
 | |
1Q92
 
 | | Crystal structure of human mitochondrial deoxyribonucleotidase in complex with the inhibitor PMcP-U | | Descriptor: | 5(3)-deoxyribonucleotidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rinaldo-Matthis, A, Rampazzo, C, Balzarini, J, Reichard, P, Bianchi, V, Nordlund, P. | | Deposit date: | 2003-08-22 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the mitochondrial deoxyribonucleotidase in complex with two specific inhibitors
Mol.Pharmacol., 65, 2004
|
|
7LOY
 
 | | Dihydrodipicolinate synthase with pyruvate from Candidatus Liberibacter solanacearum | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase | | Authors: | Gilkes, J.M, Frampton, R.A, Board, A, Sheen, C.R, Smith, G.R, Dobson, R.C.J. | | Deposit date: | 2021-02-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dihydrodipicolinate synthase with pyruvate from the plant pathogen, Candidatus Liberibacter solanacearum
To Be Published
|
|
7LVL
 
 | | Dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine from Candidatus Liberibacter solanacearum | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, LYSINE | | Authors: | Gilkes, J.M, Frampton, R.A, Board, A.J, Sheen, C.R, Smith, G.R, Dobson, R.C.J.D. | | Deposit date: | 2021-02-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine from Candidatus Liberibacter solanacearum
To Be Published
|
|
7BH4
 
 | | XFEL structure of apo CTX-M-15 after mixing for 0.7 sec with ertapenem using a piezoelectric injector (PolyPico) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH6
 
 | | Room temperature, serial X-ray structure of CTX-M-15 collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHN
 
 | |
7BHK
 
 | |
