6R54
 
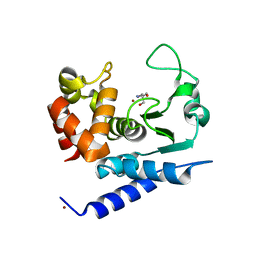 | | Crystal structure of PPEP-1(E184A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICKEL (II) ION, Pro-Pro endopeptidase, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.417 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6B2G
 
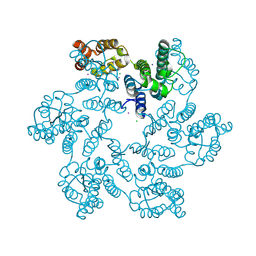 | | P38A mutant of HIV-1 capsid protein | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6R55
 
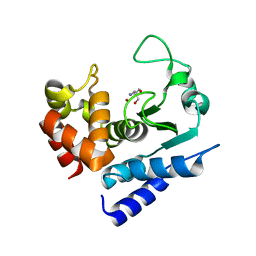 | | Crystal structure of PPEP-1(E184K) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
5SY9
 
 | |
6B2H
 
 | |
5YRS
 
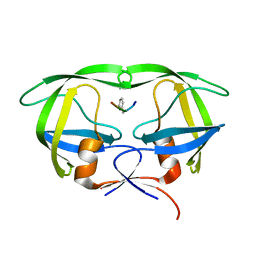 | | X-ray Snapshot of HIV-1 Protease in Action: Observation of Tetrahedral Intermediate and Its SIHB with Catalytic Aspartate | | Descriptor: | PROTEASE, RT-RH oligopeprtide | | Authors: | Das, A, Mahale, S, Prashar, V, Bihani, S, Ferrer, J.-L, Hosur, M.V. | | Deposit date: | 2017-11-10 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate.
J. Am. Chem. Soc., 132, 2010
|
|
4I0G
 
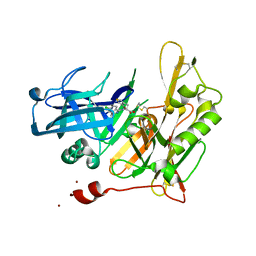 | | Design and Synthesis of Thiophene Dihydroisoquinolins as Novel BACE-1 Inhibitors | | Descriptor: | 3-(4-bromothiophen-3-yl)-N-(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)-L-alanine, Beta-secretase 1, ZINC ION | | Authors: | Yao, N, Brecht, E. | | Deposit date: | 2012-11-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HVP
 
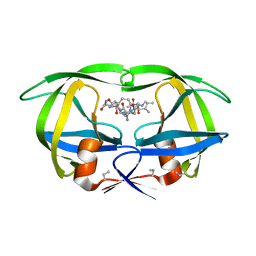 | | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 Angstroms resolution | | Descriptor: | HIV-1 PROTEASE, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide | | Authors: | Miller, M, Schneider, J, Sathyanarayana, B.K, Toth, M.V, Marshall, G.R, Clawson, L, Selk, L, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1989-08-08 | | Release date: | 1990-04-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution.
Science, 246, 1989
|
|
6KZZ
 
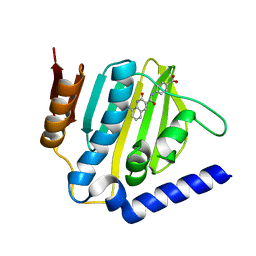 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6R5A
 
 | | Crystal structure of PPEP-1(W103F) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
3LTH
 
 | | E. cloacae MurA dead-end complex with UNAG and fosfomycin | | Descriptor: | UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, [(1R)-1-hydroxypropyl]phosphonic acid | | Authors: | Schonbrunn, E. | | Deposit date: | 2010-02-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) .
Biochemistry, 49, 2010
|
|
6IMV
 
 | | The complex structure of endo-beta-1,2-glucanase from Talaromyces funiculosus with sophorose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
5CR5
 
 | |
4YL1
 
 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 5-(4-tert-butylphenyl)-1-[4-(propan-2-yloxy)phenyl]-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
6Q92
 
 | | Crystal structure of human Arginase-1 at pH 7.0 in complex with ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase-1, MANGANESE (II) ION, ... | | Authors: | Grobben, Y, Uitdehaag, J.C.M, Zaman, G.J.R. | | Deposit date: | 2018-12-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158.
J Struct Biol X, 4, 2020
|
|
5J8O
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2R)-1-({3-bromo-4-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]phenyl}methyl)piperidine-2-carboxylic acid, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
5HVP
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF A COMPLEX BETWEEN HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE AND ACETYL-PEPSTATIN AT 2.0-ANGSTROMS RESOLUTION | | Descriptor: | ACETYL-*PEPSTATIN, CHLORIDE ION, HIV-1 PROTEASE | | Authors: | Fitzgerald, P.M.D, Mckeever, B.M, Vanmiddlesworth, J.F, Springer, J.P. | | Deposit date: | 1990-04-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of a complex between human immunodeficiency virus type 1 protease and acetyl-pepstatin at 2.0-A resolution.
J.Biol.Chem., 265, 1990
|
|
5CSO
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleoside, cytidine at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
4I2P
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with rilpivirine (TMC278) based analogue | | Descriptor: | (2E)-3-[4-({6-[(4-methoxyphenyl)amino]-7H-purin-2-yl}amino)-3,5-dimethylphenyl]prop-2-enenitrile, Gag-Pol polyprotein | | Authors: | Patel, D, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2012-11-22 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2964 Å) | | Cite: | A comparison of the ability of rilpivirine (TMC278) and selected analogues to inhibit clinically relevant HIV-1 reverse transcriptase mutants.
Retrovirology, 9, 2012
|
|
4I11
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates. | | Descriptor: | Beta-secretase 1, N-(3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)-L-phenylalanine, ZINC ION | | Authors: | Bowers, B, Xu, Y, Yuan, S, Probst, G.D, Hom, R.K, Chan, W, Konradi, A.W, Sham, H.L, Zhu, Y.L, Beroza, P, Pan, H, Brecht, E, Yao, N, Lougheed, J, Artis, D.R, Tam, D, Bova, M. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6FGY
 
 | | Crystal Structure of Human BACE-1 in Complex with amino-1,4-oxazine compound 4 | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R})-5-azanyl-3-methyl-2,6-dihydro-1,4-oxazin-3-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.-M, Bourgier, E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of amino-1,4-oxazines as potent BACE-1 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
9D3D
 
 | | Cryo-EM structure of PGT145 R100aS Fab bound to HIV-1 BG505 DS-SOSIP.664 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP glycoprotein gp41, ... | | Authors: | Hodges, S, Morano, N.C, Shapiro, L, Kwong, P.D, Gorman, J. | | Deposit date: | 2024-08-09 | | Release date: | 2024-12-25 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage.
Cell Rep, 44, 2025
|
|
9D1W
 
 | | Cryo-EM structure of PGDM1400 Fab bound to HIV-1 BG505 DS-SOSIP.664 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP glycoprotein gp41, ... | | Authors: | Kanai, T, Morano, N.C, Shapiro, L, Kwong, P.D, Gorman, J. | | Deposit date: | 2024-08-08 | | Release date: | 2024-12-25 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage.
Cell Rep, 44, 2025
|
|
5D24
 
 | | First bromodomain of BRD4 bound to inhibitor XD26 | | Descriptor: | 1,2-ETHANEDIOL, 4-acetyl-N-[3-(2-amino-2-oxoethoxy)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M, Gerhardt, S. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
9BIO
 
 | |
