6UDI
 
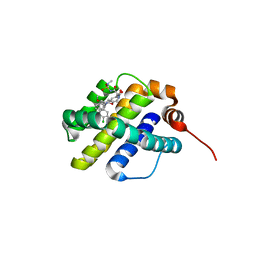 | | X-ray co-crystal structure of compound 20 with Mcl-1 | | Descriptor: | (4S,7aR,9aR,10S,11E,18R)-6'-chloro-N-(dimethylsulfamoyl)-18-hydroxy-10-methoxy-15-methyl-16-oxo-3',4',7,7a,8,9,9a,10,13,14,15,16,17,18-tetradecahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)cyclobuta[n][1,4]oxazepino[4,3-a][1,8]diazacyclohexadecine-4,1'-naphthalene]-18-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X, Whittington, D. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
6EGD
 
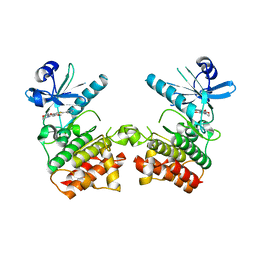 | |
3VBY
 
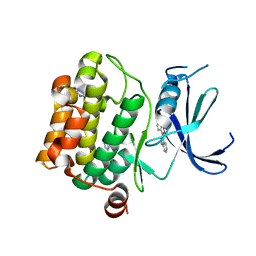 | |
4M7I
 
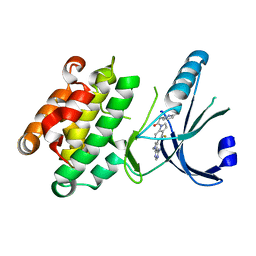 | | Crystal Structure of GSK6157 Bound to PERK (R587-R1092, delete A660-T867) at 2.34A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-4-fluoro-1H-indol-1-yl]-2-(6-methylpyridin-2-yl)ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery of 5-{4-fluoro-1-[(6-methyl-2-pyridinyl)acetyl]-2,3-dihydro-1H-indol-5-yl}-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2656157), a Potent and Selective PERK Inhibitor Selected for Preclinical Development
To be Published
|
|
6EQM
 
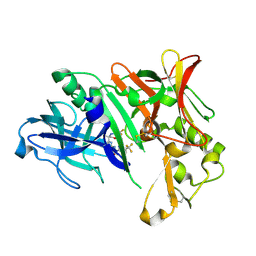 | | Crystal Structure of Human BACE-1 in Complex with CNP520 | | Descriptor: | Beta-secretase 1, ~{N}-[6-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-5-fluoranyl-pyridin-2-yl]-3-chloranyl-5-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Rondeau, J.-M, Wirth, E. | | Deposit date: | 2017-10-13 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The BACE-1 inhibitor CNP520 for prevention trials in Alzheimer's disease.
EMBO Mol Med, 10, 2018
|
|
6E8P
 
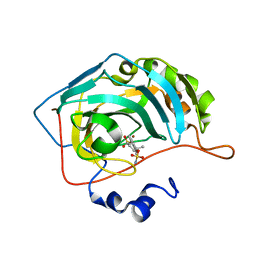 | | CA IX mimic Complexed with Steroidal Sulfamate Compound STX 49 | | Descriptor: | (14beta,17beta)-estra-1(10),2,4,6,8-pentaene-3,17-diyl disulfamate, Carbonic anhydrase 2, ZINC ION | | Authors: | Andring, J.T, Mckenna, R. | | Deposit date: | 2018-07-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3,17 beta-Bis-sulfamoyloxy-2-methoxyestra-1,3,5(10)-triene and Nonsteroidal Sulfamate Derivatives Inhibit Carbonic Anhydrase IX: Structure-Activity Optimization for Isoform Selectivity.
J. Med. Chem., 62, 2019
|
|
6SJJ
 
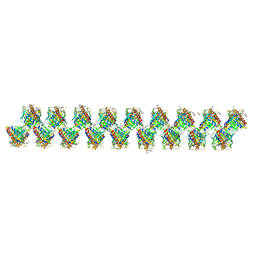 | | A new modulated crystal structure of ANS complex of St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, CITRATE ANION, ... | | Authors: | Smietanska, J, Sliwiak, J, Gilski, M, Dauter, Z, Strzalka, R, Wolny, J, Jaskolski, M. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new modulated crystal structure of the ANS complex of the St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3DS5
 
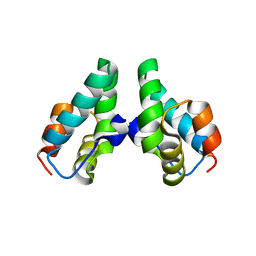 | | HIV-1 capsid C-terminal domain mutant (N183A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
6F3L
 
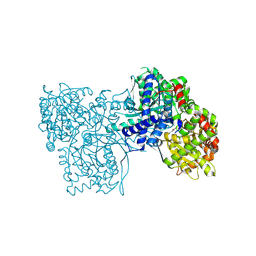 | | The crystal structure of Glycogen Phosphorylase in complex with 10b | | Descriptor: | 6-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-1~{H}-1,2,4-triazol-3-yl]naphthalene-2-carboxylic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Barkas, T.A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
5HA1
 
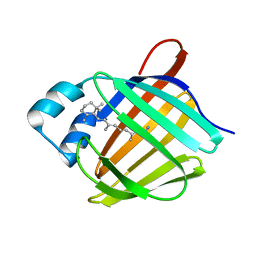 | | Crystal structure of human cellular retinol binding protein 1 in complex with retinylamine | | Descriptor: | (2~{E},4~{E},6~{E},8~{E})-3,7-dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraen-1-amine, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
3UZC
 
 | | Thermostabilised Adenosine A2A receptor in complex with 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol | | Descriptor: | 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol, Adenosine A2A Receptor | | Authors: | Congreve, M, Andrews, S.P, Dore, A.S, Hollenstein, K, Hurrell, E, Langmead, C.J, Mason, J.S, Ng, I.W, Zhukov, A, Weir, M, Marshall, F.H. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.341 Å) | | Cite: | Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
J.Med.Chem., 55, 2012
|
|
7JYR
 
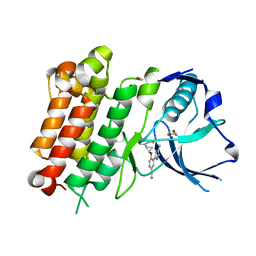 | | hALK in complex with 1-[(1R,2R)-1-(2,4-difluorophenyl)-2-[2-(5-methyl-1H-pyrazol-3-yl)-4-(trifluoromethyl)phenoxy]cyclopropyl]methanamine | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(1R,2R)-2-(2,4-difluorophenyl)cyclopropyl]oxy}-3-(5-methyl-1H-pyrazol-3-yl)benzonitrile, ALK tyrosine kinase receptor | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
6U64
 
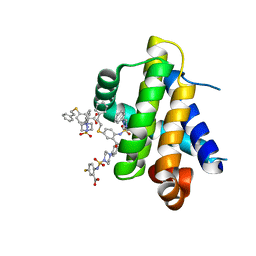 | | Mcl-1 bound to compound 17 | | Descriptor: | 5-[(2-phenylethyl)sulfanyl]-2-{[(4-phenylpiperazin-1-yl)sulfonyl]amino}benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6U67
 
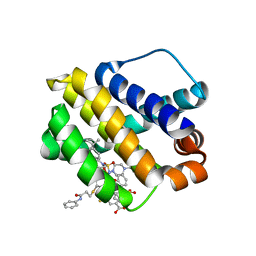 | | Mcl-1 bound to compound 24 | | Descriptor: | 2-({[4-(4-tert-butylphenyl)piperazin-1-yl]sulfonyl}amino)-5-{[3-oxo-3-(phenylamino)propyl]sulfanyl}benzoic acid, BIPHENYL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6UAX
 
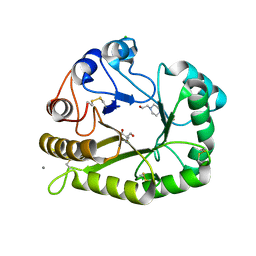 | | Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Sorangium cellulosum (ScGH128_II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Santos, C.R, Costa, P.A.C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Vieira, P.S, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6FAR
 
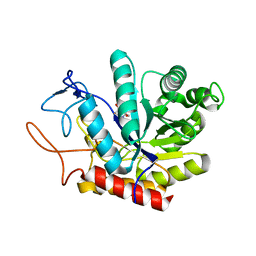 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Glycosyl hydrolase family 71, alpha-D-mannopyranose | | Authors: | Fernandes, P.Z, Petricevic, M, Sobala, L.F, Davies, G.J, Williams, S.J. | | Deposit date: | 2017-12-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Exploration of Strategies for Mechanism-Based Inhibitor Design for Family GH99 endo-alpha-1,2-Mannanases.
Chemistry, 24, 2018
|
|
5A56
 
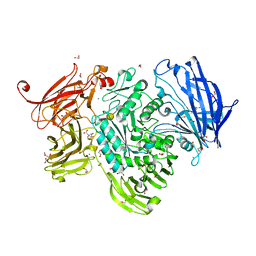 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with 1-O-methyl-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights into Its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A7G
 
 | | Comparison of the structure and activity of glycosylated and aglycosylated Human Carboxylesterase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LIVER CARBOXYLESTERASE 1 | | Authors: | Arena de Souza, V, Scott, D.J, Charlton, M, Walsh, M.A, Owen, R.J. | | Deposit date: | 2015-07-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison of the Structure and Activity of Glycosylated and Aglycosylated Human Carboxylesterase 1.
Plos One, 10, 2015
|
|
3GD9
 
 | | Crystal structure of laminaripentaose-producing beta-1,3-glucanase in complex with laminaritetraose | | Descriptor: | Laminaripentaose-producing beta-1,3-guluase (LPHase), beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Wu, H.M, Hsu, M.T, Liu, S.W, Lai, C.C, Li, Y.K, Wang, W.C. | | Deposit date: | 2009-02-23 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase.
J.Biol.Chem., 284, 2009
|
|
2UY3
 
 | |
5AAV
 
 | | Optimization of a novel binding motif to to (E)-3-(3,5-difluoro-4-((1R,3R)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H- pyrido(3,4-b)indol-1-yl)phenyl)acrylic acid (AZD9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist | | Descriptor: | (2E)-3-{4-[(1E)-1,2-DIPHENYLBUT-1-ENYL]PHENYL}ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Bradbury, R.H, de Almeida, C, Andrews, D.M, Ballard, P, Buttar, D, Callis, R.J, Currie, G.S, Curwen, J.O, Davies, C.D, de Savi, C, Donald, C.S, Feron, L.J.L, Glossop, S.C, Hayter, B.R, Karoutchi, G, Lamont, S.G, MacFaul, P, Moss, T, Pearson, S.E, Rabow, A.A, Tonge, M, Walker, G.E, Weir, H.M, Wilson, Z. | | Deposit date: | 2015-07-29 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
3VW6
 
 | | Crystal structure of human apoptosis signal-regulating kinase 1 (ASK1) with imidazopyridine inhibitor | | Descriptor: | 4-tert-butyl-N-[6-(1H-imidazol-1-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Terao, Y, Suzuki, H, Yoshikawa, M, Yashiro, H, Takekawa, S, Fujitani, Y, Okada, K, Inoue, Y, Yamamoto, Y, Nakagawa, H, Yao, S, Kawamoto, T, Uchikawa, O. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and biological evaluation of imidazo[1,2-a]pyridines as novel and potent ASK1 inhibitors.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
5HOA
 
 | | Crystal structure of c-Met L1195V in complex with SAR125844 | | Descriptor: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Marquette, J.-P. | | Deposit date: | 2016-01-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5E1D
 
 | | NTMT1 in complex with YPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E1O
 
 | | Crystal structure of NTMT1 in complex with RPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
