8UJA
 
 | |
7JL0
 
 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
1SLW
 
 | |
3CSH
 
 | |
1Q55
 
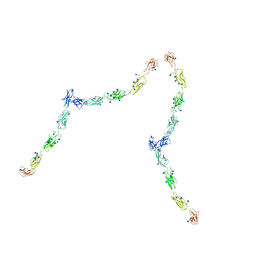 | | W-shaped trans interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
2OOL
 
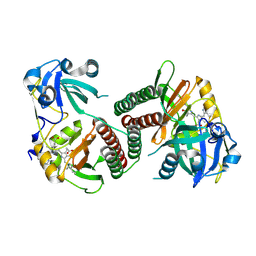 | | Crystal structure of the chromophore-binding domain of an unusual bacteriophytochrome RpBphP3 from R. palustris | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Sensor protein | | Authors: | Yang, X, Stojkovic, E.A, Kuk, J, Moffat, K. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5O31
 
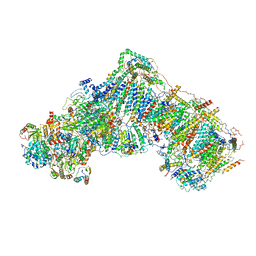 | | Mitochondrial complex I in the deactive state | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Blaza, J.N, Vinothkumar, K.R, Hirst, J. | | Deposit date: | 2017-05-23 | | Release date: | 2018-01-17 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structure of the Deactive State of Mammalian Respiratory Complex I.
Structure, 26, 2018
|
|
4LAD
 
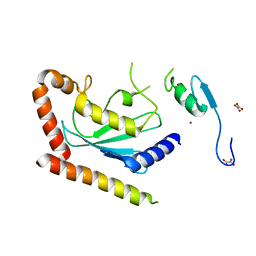 | | Crystal Structure of the Ube2g2:RING-G2BR complex | | Descriptor: | E3 ubiquitin-protein ligase AMFR, OXALATE ION, Ubiquitin-conjugating enzyme E2 G2, ... | | Authors: | Liang, Y.-H, Li, J, Das, R, Byrd, R.A, Ji, X. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
3ZYS
 
 | | Human dynamin 1 deltaPRD polymer stabilized with GMPPCP | | Descriptor: | DYNAMIN-1, INTERFERON-INDUCED GTP-BINDING PROTEIN MX1 | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
4BO2
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(1-ethylbenzimidazol-2- yl)-3-(2-methoxyphenyl)urea at 1.9A resolution | | Descriptor: | 1-(1-ethylbenzimidazol-2-yl)-3-(2-methoxyphenyl)urea, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO5
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2-chlorophenyl)-4- pyrrol-1-yl-1,3,5-triazin-2-amine at 2.6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2-chlorophenyl)-4-pyrrol-1-yl-1,3,5-triazin-2-amine, NICKEL (II) ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
1Q5B
 
 | | lambda-shaped TRANS and CIS interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
4BO7
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2,3-dihydro-1H-inden- 5-yl)tetrazolo(1,5-b)pyridazin-6-amine at 2.6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2,3-dihydro-1H-inden-5-yl)tetrazolo[1,5-b]pyridazin-6-amine | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO6
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 2,3-dihydroindol-1-yl-(2- thiophen-3-yl-1,3-thiazol-4-yl)methanone at 2.8A resolution | | Descriptor: | 2,3-dihydroindol-1-yl-(2-thiophen-3-yl-1,3-thiazol-4-yl)methanone, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO8
 
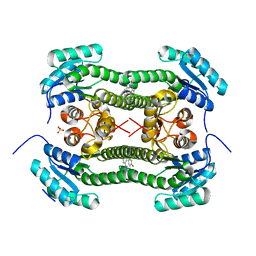 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(2-amino-4- phenylimidazol-1-yl)-3-(2-fluorophenyl)urea at 2.7A resolution | | Descriptor: | 1-(2-AMINO-4-PHENYLIMIDAZOL-1-YL)-3-(2-FLUOROPHENYL)UREA, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, SULFATE ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNT
 
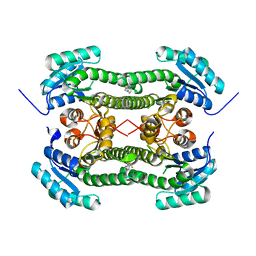 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 2-(trifluoromethyl)-1H- benzimidazole at 2.3A resolution | | Descriptor: | 2-(trifluoromethyl)-1H-benzimidazole, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNY
 
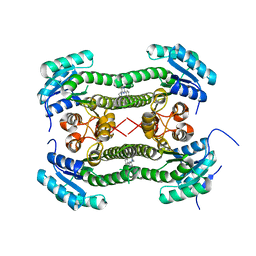 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 4-(2-phenylthieno(3,2-d) pyrimidin-4-yl)morpholine at 1.8A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 4-(2-phenylthieno[3,2-d]pyrimidin-4-yl)morpholine | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
8UI2
 
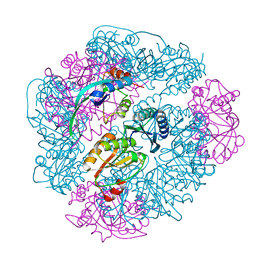 | | T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml28-redesigned-CutA-fold, T33-ml28-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-09 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UKM
 
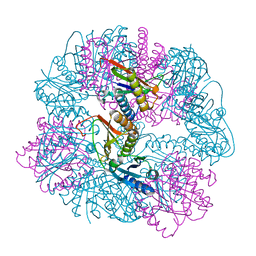 | | T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml30-redesigned-4-OT-fold, T33-ml30-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-14 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
3CSI
 
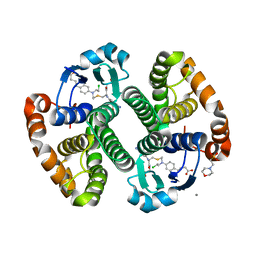 | |
1ZR4
 
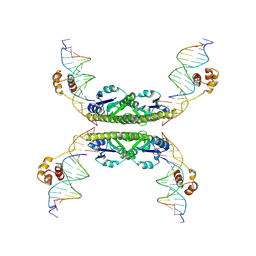 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-19 | | Release date: | 2005-08-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
1Y8D
 
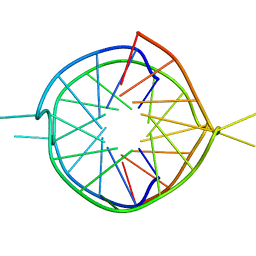 | | Dimeric parallel-stranded tetraplex with 3+1 5' G-tetrad interface, single-residue chain reversal loops and GAG triad in the context of A(GGGG) pentad | | Descriptor: | 5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*GP*T)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Ma, J.-B, Faure, A, Andreola, M.-L, Patel, D.J. | | Deposit date: | 2004-12-11 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1P1V
 
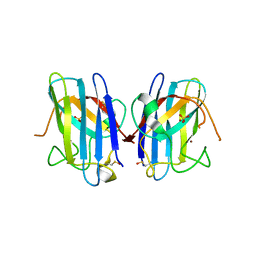 | | Crystal Structure of FALS-associated human Copper-Zinc Superoxide Dismutase (CuZnSOD) Mutant D125H to 1.4A | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Malek, K, Rodriguez, J.A, Doucette, P.A, Taylor, A.B, Hayward, L.J, Cabelli, D.E, Valentine, J.S, Hart, P.J. | | Deposit date: | 2003-04-14 | | Release date: | 2003-08-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Alternative Mechanism of Bicarbonate-mediated Peroxidation by Copper-Zinc Superoxide Dismutase: RATES ENHANCED VIA PROPOSED ENZYME-ASSOCIATED PEROXYCARBONATE INTERMEDIATE
J.Biol.Chem., 278, 2003
|
|
3D87
 
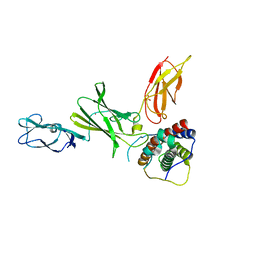 | | Crystal structure of Interleukin-23 | | Descriptor: | Interleukin-12 subunit p40, Interleukin-23 subunit p19, PHOSPHATE ION, ... | | Authors: | Beyer, B.M, Ingram, R, Ramanathan, L, Reichert, P, Le, H, Madison, V. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody
J.Mol.Biol., 382, 2008
|
|
1ZR2
 
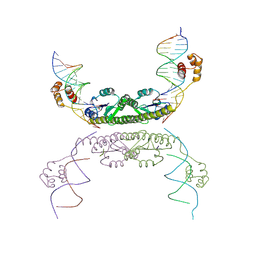 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently Linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-18 | | Release date: | 2005-08-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
