5LJI
 
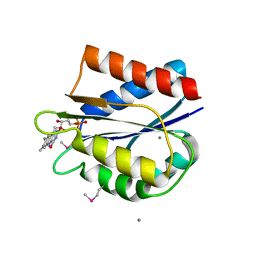 | | Streptococcus pneumonia TIGR4 flavodoxin: structural and biophysical characterization of a novel drug target | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Rodriguez-Cardenas, A, Rojas, A.L, Velazquez-Campoy, A, Hurtado-Guerrero, R, Sancho, J. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Streptococcus pneumoniae TIGR4 Flavodoxin: Structural and Biophysical Characterization of a Novel Drug Target.
Plos One, 11, 2016
|
|
4KO2
 
 | | Low X-ray dose structure of H2-activated anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, GLYCEROL, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem. Commun. (Camb.), 49, 2013
|
|
4KU2
 
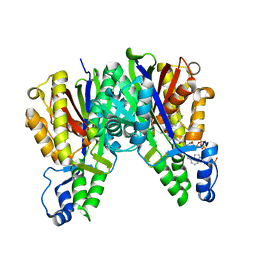 | |
3GS6
 
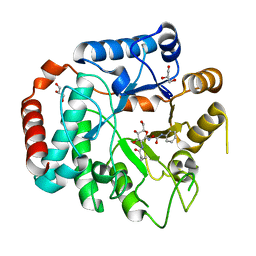 | | Vibrio Cholerea family 3 glycoside hydrolase (NagZ)in complex with N-butyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-3-(BUTANOYLAMINO)-4,5-DIHYDROXY-6-(HYDROXYMETHYL)OXAN-2-YLIDENE]AMINO] N-PHENYLCARBAMATE | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
3GSM
 
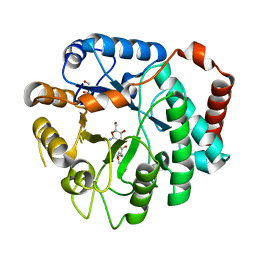 | | Vibrio cholerae family 3 glycoside hydrolase (NagZ) bound to N-Valeryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-(pentanoylamino)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
4KTI
 
 | | Crystal Structure of C143A Xathomonas campestris OleA | | Descriptor: | 3-oxoacyl-[ACP] synthase III | | Authors: | Goblirsch, B.R. | | Deposit date: | 2013-05-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Substrate Trapping in Crystals of the Thiolase OleA Identifies Three Channels That Enable Long Chain Olefin Biosynthesis.
J.Biol.Chem., 291, 2016
|
|
2QS3
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP316 at 1.76 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-5-phenylthiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | ACET is a highly potent and specific kainate receptor antagonist: characterisation and effects on hippocampal mossy fibre function.
Neuropharmacology, 56, 2009
|
|
4KU5
 
 | |
2R9X
 
 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | Descriptor: | 2-[(1R)-2-carboxy-1-(naphthalen-1-ylmethyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindole-5-carboxylic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Babaoglu, K, Shoichet, B.K. | | Deposit date: | 2007-09-13 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
2R9W
 
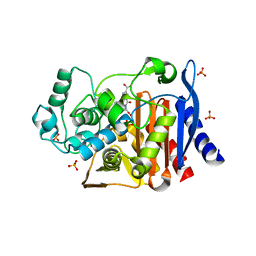 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | Descriptor: | 2-[(1R)-1-carboxy-2-naphthalen-1-ylethyl]-1,3-dioxo-2,3-dihydro-1H-isoindole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Babaoglu, K, Shoichet, B.K. | | Deposit date: | 2007-09-13 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
4KU3
 
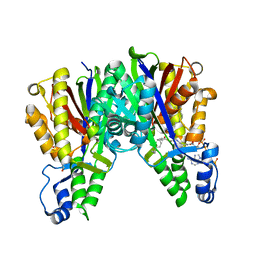 | |
1CC1
 
 | | CRYSTAL STRUCTURE OF A REDUCED, ACTIVE FORM OF THE NI-FE-SE HYDROGENASE FROM DESULFOMICROBIUM BACULATUM | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROGENASE (LARGE SUBUNIT), ... | | Authors: | Garcin, E, Vernede, X, Hatchikian, E.C, Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1999-03-03 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center
Structure Fold.Des., 7, 1999
|
|
1P0I
 
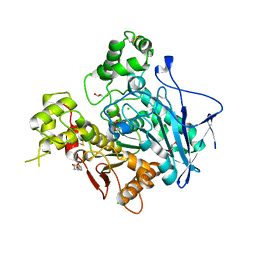 | | Crystal structure of human butyryl cholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
1CLQ
 
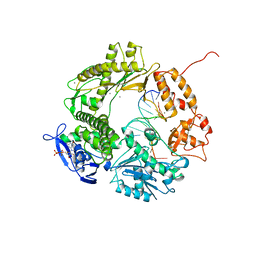 | |
7STS
 
 | | Crystal Structure of Human Fab S24-1379 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | Fab S24-1379, heavy chain, light chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7SUE
 
 | | Crystal Structure of Human Fab S24-188 in the complex with the N-teminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | Nucleoprotein, S24-188 Fab Heavy chain, S24-188 Fab Light chain | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
4HVK
 
 | | Crystal structure and functional studies of an unusual L-cysteine desulfurase from Archaeoglobus fulgidus. | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yamanaka, Y, Zeppieri, L, Nicolet, Y, Marinoni, E.N, de Oliveira, J.S, Masafumi, O, Dean, D.R, Fontecilla-Camps, J.C. | | Deposit date: | 2012-11-06 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure and functional studies of an unusual L-cysteine desulfurase from Archaeoglobus fulgidus.
Dalton Trans, 42, 2013
|
|
4I4T
 
 | | Crystal structure of tubulin-RB3-TTL-Zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism of Action of Microtubule-Stabilizing Anticancer Agents.
Science, 339, 2013
|
|
8D1U
 
 | | E. coli beta-ketoacyl-[acyl carrier protein] synthase III (FabH) with an acetylated cysteine and in complex with oxa(dethia)-Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, CHLORIDE ION, oxa(dethia)-CoA | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-ketoacylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(dethia)CoA.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
1QDD
 
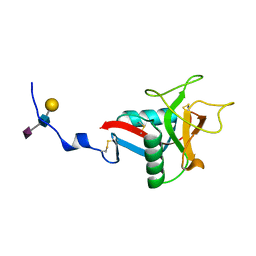 | | CRYSTAL STRUCTURE OF HUMAN LITHOSTATHINE TO 1.3 A RESOLUTION | | Descriptor: | LITHOSTATHINE, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Gerbaud, V, Pignol, D, Loret, E, Bertrand, J.A, Berland, Y, Fontecilla-Camps, J.C, Canselier, J.P, Gabas, N, Verdier, J.M. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of calcite crystal growth inhibition by the N-terminal undecapeptide of lithostathine.
J.Biol.Chem., 275, 2000
|
|
1MRW
 
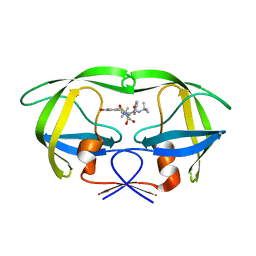 | | Structure of HIV protease (Mutant Q7K L33I L63I) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
7CFD
 
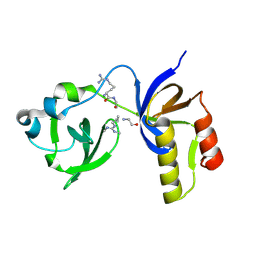 | | Drosophila melanogaster Krimper eTud2-AubR15me2 complex | | Descriptor: | FI20010p1, Protein aubergine | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFB
 
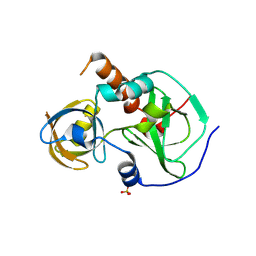 | | Drosophila melanogaster Krimper eTud1 apo structure | | Descriptor: | FI20010p1, SULFATE ION | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFC
 
 | | Drosophila melanogaster Krimper eTud1-Ago3 complex | | Descriptor: | FI20010p1, Protein argonaute-3 | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
