5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
1Z6V
 
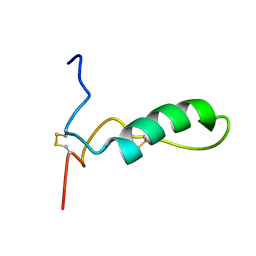 | | Human lactoferricin | | Descriptor: | Lactotransferrin | | Authors: | Hunter, H.N, Demcoe, A.R, Jenssen, H, Gutteberg, T.J, Vogel, H.J. | | Deposit date: | 2005-03-23 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Human lactoferricin is partially folded in aqueous solution and is better stabilized in a membrane mimetic solvent
ANTIMICROB.AGENTS CHEMOTHER., 49, 2005
|
|
6K31
 
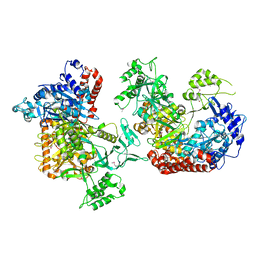 | |
3PL9
 
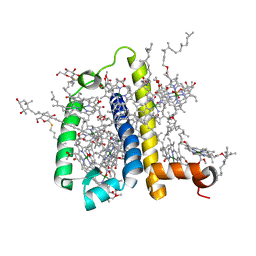 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | Deposit date: | 2010-11-14 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1Z6W
 
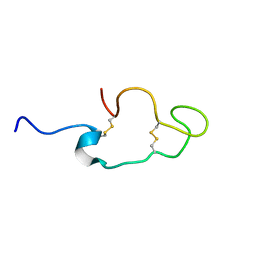 | | Human Lactoferricin | | Descriptor: | Lactotransferrin | | Authors: | Hunter, H.N, Demcoe, A.R, Jenssen, H, Gutteberg, T.J, Vogel, H.J. | | Deposit date: | 2005-03-23 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Human Lactoferricin Is Partially Folded in Aqueous Solution and Is Better Stabilized in a Membrane Mimetic Solvent
ANTIMICROB.AGENTS CHEMOTHER., 49, 2005
|
|
2LHO
 
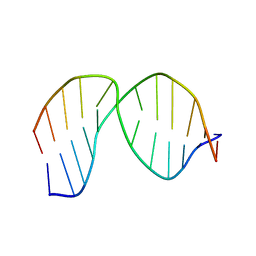 | | Solution Structure of a DNA duplex Containing an Unnatural, Hydrophobic Base Pair | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*TP*CP*(LHO)P*TP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*AP*(MM7)P*GP*AP*AP*AP*CP*G)-3') | | Authors: | Malyshev, D.A, Pfaff, D.A, Ippoliti, S.L, Hwang, G.T, Dwyer, T.J, Romesberg, F.E. | | Deposit date: | 2011-08-12 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure, mechanism of replication, and optimization of an unnatural base pair.
Chemistry, 16, 2010
|
|
2JTF
 
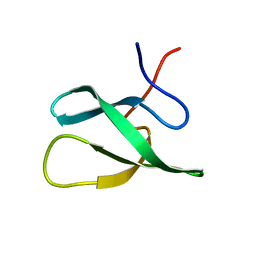 | | Solution Structure of the PHF20L1 MBT domain | | Descriptor: | PHD finger protein 20-like 1 | | Authors: | Brockmann, C, Iberg, A.N, Rehbein, K, Diehl, A, Bedford, M.T, Oschkinat, H. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of Histone H4K20 Methyllysine Recognition by the MBT Domain of PHF20L1
To be Published
|
|
2JOU
 
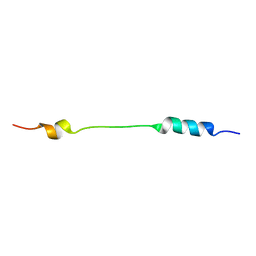 | | NMR structure of Mini-B, an N-terminal- C-terminal construct from human Surfactant Protein-B (SP-B), in Hexafluoroisopropanol (HFIP) | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Booth, V, Sarker, M, Keough, K.M.W, Waring, A.J, Walther, F.J. | | Deposit date: | 2007-03-26 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of mini-B, a functional fragment of surfactant protein B, in detergent micelles
Biochemistry, 46, 2007
|
|
2JSG
 
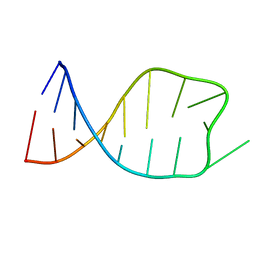 | | NMR solution structure of the anticodon of E.coli TRNA-VAL3 with 1 modification (M6A37) | | Descriptor: | 5'-R(*CP*CP*UP*CP*CP*CP*UP*UP*AP*CP*(6MZ)P*AP*GP*GP*AP*GP*G)-3' | | Authors: | Vendeix, F.A.P, Dziergowska, A, Gustilo, E.M, Graham, W.D, Sproat, B, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2007-07-04 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Wobble-Position Modifications Pre-structure tRNA's Anticodon for Ribosome-Mediated Codon Binding
To be Published
|
|
2JVL
 
 | | NMR structure of the C-terminal domain of MBF1 of Trichoderma reesei | | Descriptor: | TrMBF1 | | Authors: | Kopke Salinas, R, Tomaselli, S, Camilo, C.M, Valencia, E.Y, Farah, C.S, El-Dorry, H, Chambergo, F.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of multiprotein bridging factor 1 (MBF1) of Trichoderma reesei.
Proteins, 75, 2009
|
|
2JSS
 
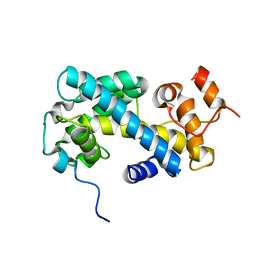 | | NMR structure of chaperone Chz1 complexed with histone H2A.Z-H2B | | Descriptor: | Chimera of Histone H2B.1 and Histone H2A.Z, Uncharacterized protein YER030W | | Authors: | Zhou, Z, Feng, H, Hansen, D.F, Kato, H, Luk, E, Freedberg, D.I, Kay, L.E, Wu, C, Bai, Y. | | Deposit date: | 2007-07-11 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6HXQ
 
 | |
6HXK
 
 | |
6HXM
 
 | |
8HFQ
 
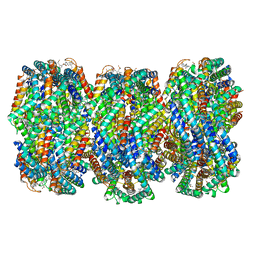 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | Authors: | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
7SAL
 
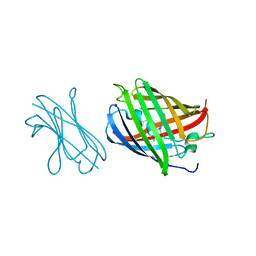 | |
7SAI
 
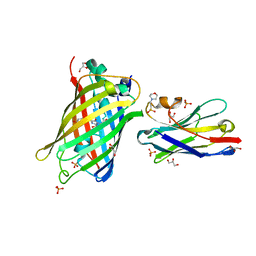 | | Crystal Structure of Lag30 Nanobody bound to eGFP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Green fluorescent protein, ... | | Authors: | Cong, A.T.Q, Schellenberg, M.J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | High-efficiency recombinant protein purification using mCherry and YFP nanobody affinity matrices.
Protein Sci., 31, 2022
|
|
7SAK
 
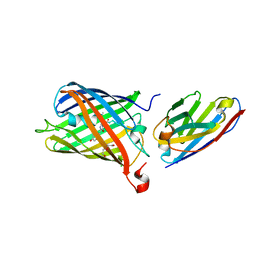 | |
7SAJ
 
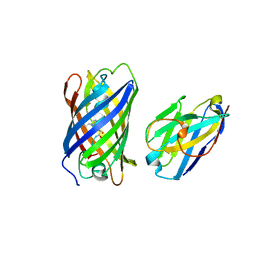 | |
8EQF
 
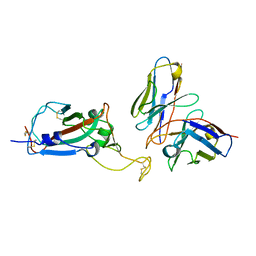 | | cryoEM structure of a broadly neutralizing anti-SARS-CoV-2 antibody STI-9167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Bajic, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | An in vitro experimental pipeline to characterize the epitope of a SARS-CoV-2 neutralizing antibody.
Mbio, 15, 2024
|
|
7KSC
 
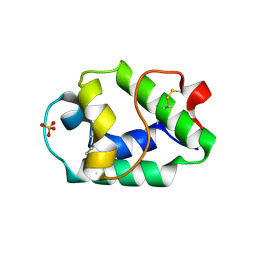 | | Crystal structure of Pun g 1.0101 | | Descriptor: | Non-specific lipid-transfer protein, SULFATE ION | | Authors: | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Tuppo, L, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2020-11-21 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
4WFN
 
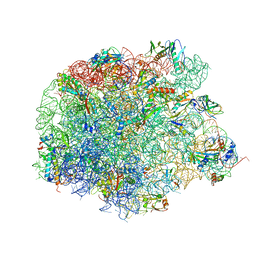 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 in complex with erythromycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-16 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
8UR9
 
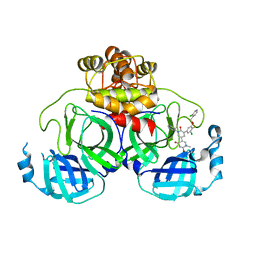 | | Crystal Structure of the SARS-CoV-2 Main Protease in Complex with Compound 61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Papini, C, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-10-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proof-of-concept studies with a computationally designed M pro inhibitor as a synergistic combination regimen alternative to Paxlovid.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
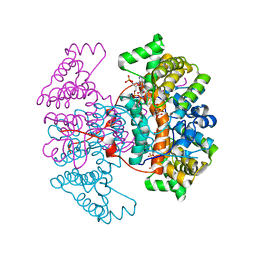
6HXH
 
 | | Structure of the human ATP citrate lyase holoenzyme in complex with citrate, coenzyme A and Mg.ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase,Human ATP citrate lyase, CITRATE ANION, ... | | Authors: | Verstraete, K, Verschueren, K. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature, 568, 2019
|
|
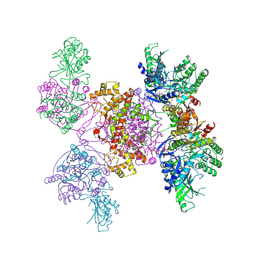
6HXI
 
 | |
