7JNM
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J36 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*GP*TP*GP*AP*CP*GP*AP*CP*GP*AP*GP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JOL
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 duplex version) containing the J24 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*GP*TP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.114 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JOK
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 duplex version) containing the J26 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JS0
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 duplex version) containing the J31 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*GP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JP6
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 duplex version) containing the J20 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*GP*AP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-08-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.087 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JRY
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 duplex version) containing the J4 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*GP*GP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JON
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 duplex version) containing the J23 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JP7
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 duplex version) containing the J16 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*GP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-08-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
1DA3
 
 | | THE CRYSTAL STRUCTURE OF THE TRIGONAL DECAMER C-G-A-T-C-G-6MEA-T-C-G: A B-DNA HELIX WITH 10.6 BASE-PAIRS PER TURN | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*AP*TP*CP*GP*(6MA)P*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Baikalov, I, Grzeskowiak, K, Yanagi, K, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-11-09 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn.
J.Mol.Biol., 231, 1993
|
|
1ONM
 
 | | Solution Structure of a DNA duplex containing A:G mismatch. d(GCTTCAGTCGT):d(ACGACGGAAGC) | | Descriptor: | 5'-D(*AP*CP*GP*AP*CP*GP*GP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*CP*AP*GP*TP*CP*GP*T)-3' | | Authors: | Sanchez, A.M, Volk, D.E, Gorenstein, D.G, Lloyd, R.S. | | Deposit date: | 2003-02-28 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Initiation of repair of A/G mismatches is modulated by sequence context
DNA REPAIR, 2, 2003
|
|
232D
 
 | |
1U76
 
 | |
5IWD
 
 | | HCMV DNA polymerase subunit UL44 complex with a small molecule | | Descriptor: | 5-methylidene-3-(methylsulfanyl)-2-benzothiophen-4(5H)-one, DNA polymerase processivity factor | | Authors: | Chen, H, Coen, D.M, Hogle, J.M, Filman, D.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A Small Covalent Allosteric Inhibitor of Human Cytomegalovirus DNA Polymerase Subunit Interactions.
ACS Infect Dis, 3, 2017
|
|
4XTJ
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl plus 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5CCW
 
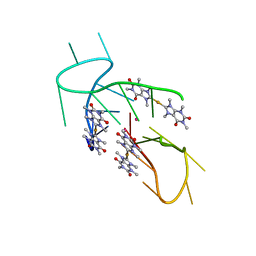 | | Structure of the complex of a human telomeric DNA with Au(caffein-2-ylidene)2 | | Descriptor: | Au(caffein-2-ylidene)2, POTASSIUM ION, human telomeric DNA | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Messori, L, Papi, F. | | Deposit date: | 2015-07-02 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Determinants for Tight and Selective Binding of a Medicinal Dicarbene Gold(I) Complex to a Telomeric DNA G-Quadruplex: a Joint ESI MS and XRD Investigation.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8ASK
 
 | | Crystal structure of d(GCCCACCACGGC) | | Descriptor: | DNA (5'-D(P*GP*CP*CP*CP*AP*CP*CP*AP*CP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*TP*GP*GP*TP*GP*GP*GP*C)-3') | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L, Rusev, R, Heroux, A, Doukov, T, Kuvandjiev, N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Structural Characterization of Alzheimer DNA Promoter Sequences from the Amyloid Precursor Gene in the Presence of Thioflavin T and Analogs
Crystals, 12, 2022
|
|
6QJS
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpiperidinoxyl (nitroxide) spin label | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*(S6M)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
3JAA
 
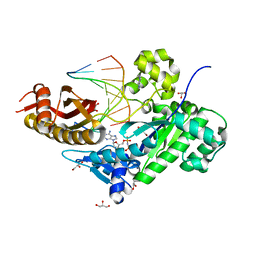 | | HUMAN DNA POLYMERASE ETA in COMPLEX WITH NORMAL DNA AND INCO NUCLEOTIDE (NRM) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3, DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Lau, W.C.Y, Li, Y, Zhang, Q, Huen, M.S.Y. | | Deposit date: | 2015-05-19 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Molecular architecture of the Ub-PCNA/Pol eta complex bound to DNA
Sci Rep, 5, 2015
|
|
3R6R
 
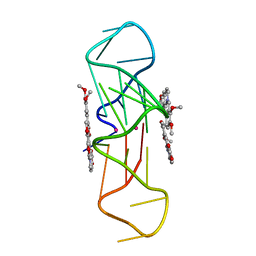 | | Structure of the complex of an intramolecular human telomeric DNA with Berberine formed in K+ solution | | Descriptor: | BERBERINE, DNA (22-mer), POTASSIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2011-03-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1.
Nucleic Acids Res., 41, 2013
|
|
1SX5
 
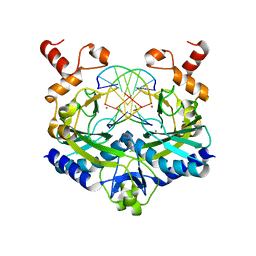 | |
1SUZ
 
 | |
1DNK
 
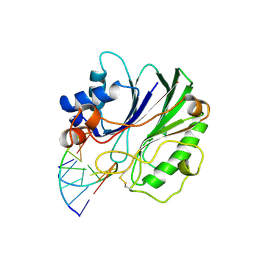 | | THE X-RAY STRUCTURE OF THE DNASE I-D(GGTATACC)2 COMPLEX AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*GP*GP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3'), ... | | Authors: | Weston, S.A, Lahm, A, Suck, D. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution.
J.Mol.Biol., 226, 1992
|
|
3M4I
 
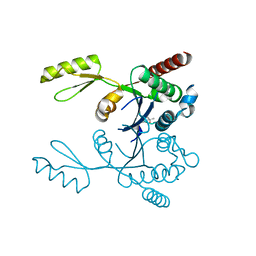 | | Crystal structure of the second part of the Mycobacterium tuberculosis DNA gyrase reaction core: the TOPRIM domain at 1.95 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B | | Authors: | Piton, J, Petrella, S, Aubry, A, Mayer, C. | | Deposit date: | 2010-03-11 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase.
Plos One, 5, 2010
|
|
2L45
 
 | | C-terminal zinc knuckle of the HIVNCp7 with DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
2L46
 
 | | C-terminal zinc finger of the HIVNCp7 with platinated DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
