9G1S
 
 | | Fragment screening of FosAKP, cryo structure in complex with fragment F2X-entry H01 | | Descriptor: | 1,2-ETHANEDIOL, Fosfomycin resistance protein, MANGANESE (II) ION, ... | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
7R7X
 
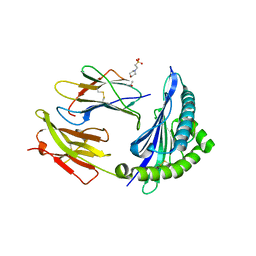 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
5MRW
 
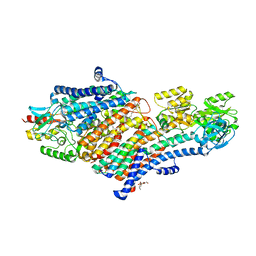 | | Structure of the KdpFABC complex | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Huang, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the potassium-importing KdpFABC membrane complex.
Nature, 546, 2017
|
|
6TBG
 
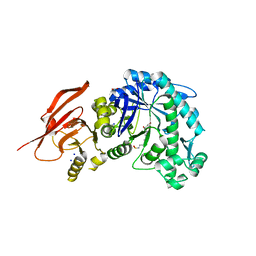 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[[(1~{R},2~{R},3~{S},4~{S},5~{S})-2-(hydroxymethyl)-3,4,5-tris(oxidanyl)cyclopentyl]amino]hexyl]naphthalene-1-sulfonamide, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
5JYB
 
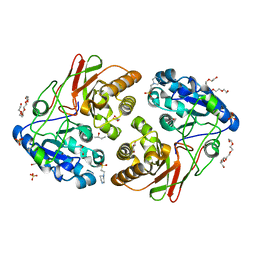 | | Crystal structure of 3 mutant of Ba3275 (S116A, E243A, H313A), the member of S66 family of serine peptidases | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nocek, B, Jedrzejczak, R, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of 3 mutant of Ba3275 (S116A, E243A, H313A), the member of S66 family of serine peptidases
To Be Published
|
|
8HVC
 
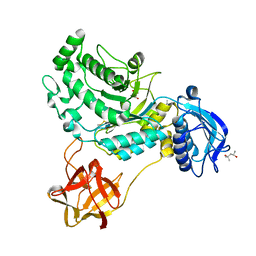 | | Crystal structure of lacto-N-biosidase StrLNBase from Streptomyces sp. strain 142, galacto-N-biose complex 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Fushinobu, S, Yamada, C, Fujio, N. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase Family 20 Lacto- N -biosidase from Soil Bacterium Streptomyces sp. Strain 142.
J Appl Glycosci (1999), 72, 2025
|
|
6HWF
 
 | | Yeast 20S proteasome beta2-G45A mutant in complex with ONX 0914 | | Descriptor: | (2~{S})-3-(4-methoxyphenyl)-~{N}-[(2~{S},3~{R})-4-methyl-3,4-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
5MXV
 
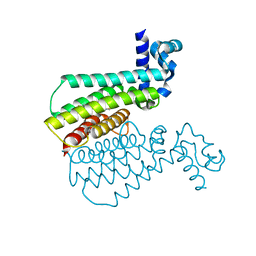 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK1107112A at 1.63A resolution | | Descriptor: | 3-chloranyl-~{N}-(4,5-dihydro-1,3-thiazol-2-yl)-6-fluoranyl-1-benzothiophene-2-carboxamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
9DQ1
 
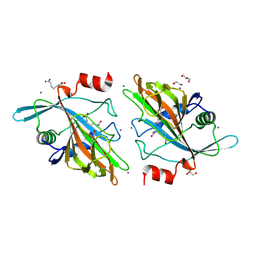 | | Crystal structure of HrmJ from Streptomyces sp. CFMR 7 (HrmJ-ssc) complexed with manganese (II), 2-oxoglutarate and 6-nitronorleucine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 6-nitro-L-norleucine, ... | | Authors: | Zheng, Y.-C, Swartz, P, Chang, W.-C. | | Deposit date: | 2024-09-23 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of a Nonheme Iron Cyclopropanase with a Homologous Hydroxylase Reveals Mechanistic Features Associated with Distinct Reaction Outcomes.
J.Am.Chem.Soc., 147, 2025
|
|
7YAS
 
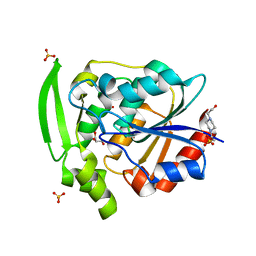 | | HYDROXYNITRILE LYASE, LOW TEMPERATURE NATIVE STRUCTURE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (HYDROXYNITRILE LYASE), ... | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
6T7G
 
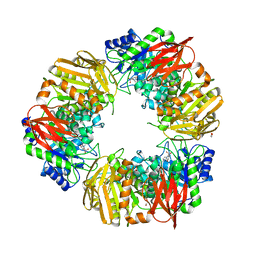 | | Bacteroides salyersiae GH164 beta-mannosidase in complex with mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Armstrong, Z, Davies, G. | | Deposit date: | 2019-10-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function ofBs164 beta-mannosidase fromBacteroides salyersiaethe founding member of glycoside hydrolase family GH164.
J.Biol.Chem., 295, 2020
|
|
6PUY
 
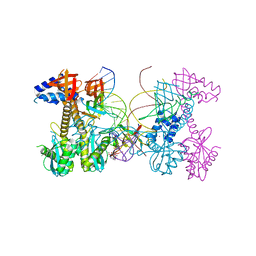 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
5M73
 
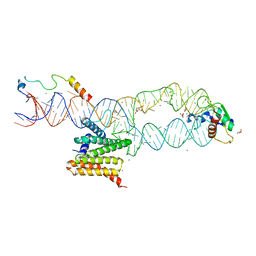 | | Structure of the human SRP S domain with SRP72 RNA-binding domain | | Descriptor: | GLYCEROL, Human gene for small cytoplasmic 7SL RNA (7L30.1), MAGNESIUM ION, ... | | Authors: | Becker, M.M.M, Wild, K, Sinning, I. | | Deposit date: | 2016-10-26 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human SRP72 complexes provide insights into SRP RNA remodeling and ribosome interaction.
Nucleic Acids Res., 45, 2017
|
|
8R2K
 
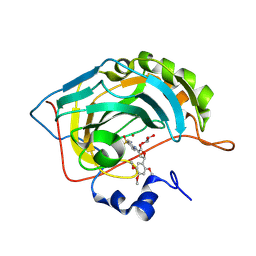 | | Human Carbonic Anhydrase II in complex with 4-((2-oxo-5-(3,4,5-trimethoxyphenyl)-2,5-dihydrofuran-3-yl)amino)benzenesulfonamide | | Descriptor: | 4-[[(2~{R})-5-oxidanylidene-2-(2,4,5-trimethoxyphenyl)-2~{H}-furan-4-yl]amino]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-11-06 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synthetic Approaches to Novel Human Carbonic Anhydrase Isoform Inhibitors Based on Pyrrol-2-one Moiety.
J.Med.Chem., 67, 2024
|
|
8DJD
 
 | |
5M8E
 
 | | Crystal structure of a GH43 arabonofuranosidase from Weissella sp. strain 142 | | Descriptor: | Alpha-N-arabinofuranosidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Linares-Pasten, J, Logan, D.T, Nordberg Karlsson, E. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures and functional studies of two GH43 arabinofuranosidases from Weissella sp. strain 142 and Lactobacillus brevis.
FEBS J., 284, 2017
|
|
7XVK
 
 | |
6HCB
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.9 A RESOLUTION. | | Descriptor: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Masternak, M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
8R0Y
 
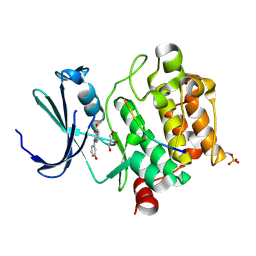 | |
7YC3
 
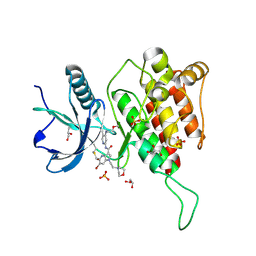 | | Crystal structure of FGFR4 kinase domain with 10t | | Descriptor: | 6-bromanyl-~{N}-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide, Fibroblast growth factor receptor 4, GLYCEROL, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
9G9U
 
 | | The structure of XynX, a NIF3 family protein from Geobacillus proteiniphilus T-6 | | Descriptor: | 1,2-ETHANEDIOL, GTP cyclohydrolase 1 type 2 homolog, ZINC ION | | Authors: | Hadad, N, Pomyalov, S, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2024-07-25 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of XynX, a NIF3 family protein from Geobacillus proteiniphilus T-6
To Be Published
|
|
6PWL
 
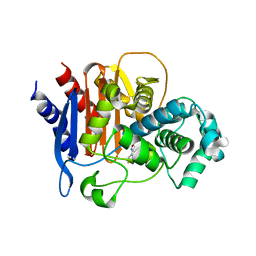 | | ADC-7 in complex with boronic acid transition state inhibitor LP06 | | Descriptor: | (7Z)-7-(2-amino-1,3-thiazol-4-yl)-1,1,3-trihydroxy-10,10-dimethyl-1,6-dioxo-2,9-dioxa-5,8-diaza-1lambda~5~-phospha-3-boraundec-7-en-11-oic acid, Beta-lactamase, GLYCINE | | Authors: | Curtis, B.C, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-07-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights into Inhibition of the Acinetobacter-Derived Cephalosporinase ADC-7 by Ceftazidime and Its Boronic Acid Transition State Analog.
Antimicrob.Agents Chemother., 64, 2020
|
|
7YC1
 
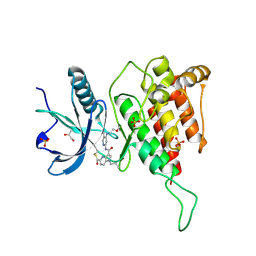 | | Crystal structure of FGFR4 kinase domain with 10d | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
8QZ7
 
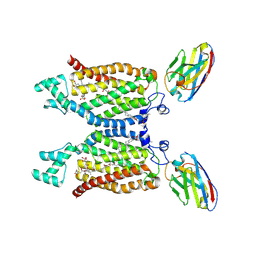 | | Structure of human ceramide synthase 6 (CerS6) in complex with N-palmitoyl fumonisin B1 | | Descriptor: | (2~{R})-2-[2-[(5~{R},6~{R},7~{S},9~{S},11~{R},16~{R},18~{S},19~{S})-6-[(3~{R})-3-carboxy-5-oxidanyl-5-oxidanylidene-pentanoyl]oxy-19-(hexadecanoylamino)-5,9-dimethyl-11,16,18-tris(oxidanyl)icosan-7-yl]oxy-2-oxidanylidene-ethyl]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6Q17
 
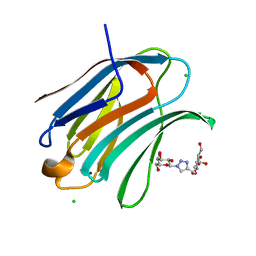 | | Crystal structure of Human galectin-3 CRD in complex with Methyl 3-O-[1-(b-D-galactopyranosyl)-1,2,3-triazol-4-yl]-methyl-b-D-galactopyranoside | | Descriptor: | CHLORIDE ION, Galectin-3, methyl 3-O-[(1-beta-D-galactopyranosyl-1H-1,2,3-triazol-4-yl)methyl]-beta-D-galactopyranoside | | Authors: | Kishor, C, Blanchard, H. | | Deposit date: | 2019-08-02 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Linear triazole-linked pseudo oligogalactosides as scaffolds for galectin inhibitor development.
Chem.Biol.Drug Des., 96, 2020
|
|
