2Q8D
 
 | | Crystal structure of JMJ2D2A in ternary complex with histone H3-K36me2 and succinate | | Descriptor: | HISTONE 3 peptide, JmjC domain-containing histone demethylation protein 3A, NICKEL (II) ION, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2Q8E
 
 | | Specificity and Mechanism of JMJD2A, a Trimethyllysine-Specific Histone Demethylase | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2Q8F
 
 | | Structure of pyruvate dehydrogenase kinase isoform 1 | | Descriptor: | POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2Q8G
 
 | | Structure of pyruvate dehydrogenase kinase isoform 1 in complex with glucose-lowering drug AZD7545 | | Descriptor: | 4-[(3-CHLORO-4-{[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]AMINO}PHENYL)SULFONYL]-N,N-DIMETHYLBENZAMIDE, POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2Q8H
 
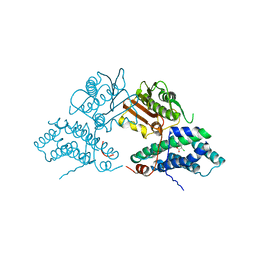 | | Structure of pyruvate dehydrogenase kinase isoform 1 in complex with dichloroacetate (DCA) | | Descriptor: | DICHLORO-ACETIC ACID, POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2Q8I
 
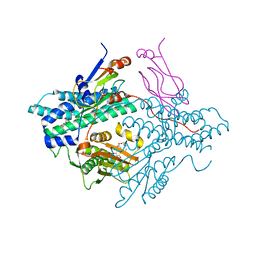 | | Pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug radicicol | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, GLYCEROL, ... | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2Q8J
 
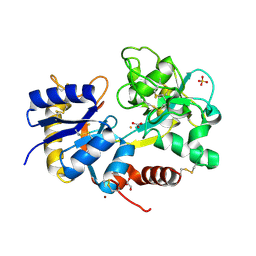 | | Crystal Structure of the complex of C-lobe of bovine lactoferrin with Mannitol and Mannose at 2.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Jain, R, Sinha, M, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of the complex of C-lobe of bovine lactoferrin with Mannitol and Mannose at 2.7 A resolution
To be Published
|
|
2Q8K
 
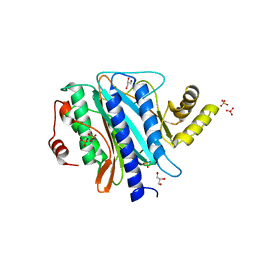 | | The crystal structure of Ebp1 | | Descriptor: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | Authors: | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
2Q8L
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Plasmodium falciparum | | Descriptor: | Orotidine-monophosphate-decarboxylase, PHOSPHATE ION | | Authors: | Liu, Y, Lau, W, Lew, J, Amani, M, Hui, R, Pai, E.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Plasmodium falciparum.
To be Published
|
|
2Q8M
 
 | | T-like Fructose-1,6-bisphosphatase from Escherichia coli with AMP, Glucose 6-phosphate, and Fructose 1,6-bisphosphate bound | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Hines, J.K, Kruesel, C.E, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Inhibited Fructose-1,6-bisphosphatase from Escherichia coli: DISTINCT ALLOSTERIC INHIBITION SITES FOR AMP AND GLUCOSE 6-PHOSPHATE AND THE CHARACTERIZATION OF A GLUCONEOGENIC SWITCH.
J.Biol.Chem., 282, 2007
|
|
2Q8N
 
 | |
2Q8O
 
 | |
2Q8P
 
 | |
2Q8Q
 
 | |
2Q8R
 
 | |
2Q8S
 
 | |
2Q8T
 
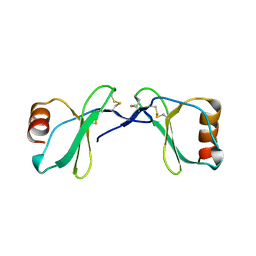 | | Crystal Structure of the CC chemokine CCL14 | | Descriptor: | CCL14 | | Authors: | Blain, K.Y. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Functional Characterization of CC Chemokine CCL14
Biochemistry, 46, 2007
|
|
2Q8U
 
 | |
2Q8V
 
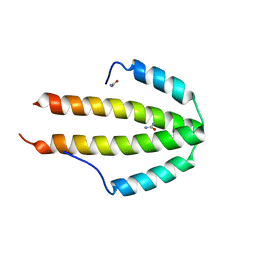 | | NblA protein from T. vulcanus crystallized with urea | | Descriptor: | NblA protein, UREA | | Authors: | Adir, N, Dines, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, Functional, and Mutational Analysis of the NblA Protein Provides Insight into Possible Modes of Interaction with the Phycobilisome
J.Biol.Chem., 283, 2008
|
|
2Q8W
 
 | |
2Q8X
 
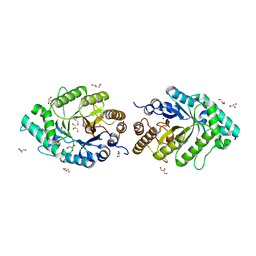 | | The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus | | Descriptor: | GLYCEROL, SODIUM ION, intra-cellular xylanase | | Authors: | Solomon, V, Teplitsky, A, Gilboa, R, Zolotnitsky, G, Golan, G, Reiland, V, Moryles, S, Shoham, Y, Shoham, G. | | Deposit date: | 2007-06-12 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2Q8Y
 
 | |
2Q8Z
 
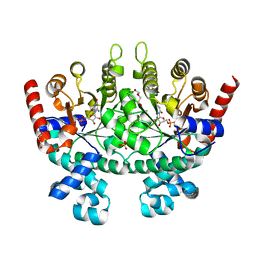 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase complexed with 6-amino-UMP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 6-AMINOURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Y, Lau, W, Bello, A.M, Kotra, L.P, Hui, R, Pai, E.F. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of C6-Uridine Derivatives Targeting Plasmodia Orotidine Monophosphate Decarboxylase.
J.Med.Chem., 51, 2008
|
|
2Q91
 
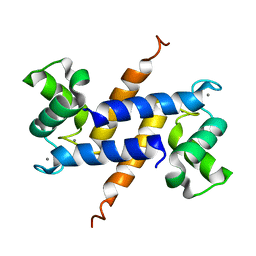 | | Structure of the Ca2+-Bound Activated Form of the S100A4 Metastasis Factor | | Descriptor: | CALCIUM ION, S100A4 Metastasis Factor | | Authors: | Malashkevich, V.N, Knight, D, Ramagopal, U.A, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2007-06-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of Ca(2+)-Bound S100A4 and Its Interaction with Peptides Derived from Nonmuscle Myosin-IIA.
Biochemistry, 47, 2008
|
|
2Q92
 
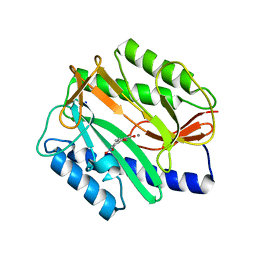 | | E. coli methionine aminopeptidase Mn-form with inhibitor B23 | | Descriptor: | 5-(2-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
