1AL3
 
 | |
1AN1
 
 | | LEECH-DERIVED TRYPTASE INHIBITOR/TRYPSIN COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPTASE INHIBITOR | | Authors: | Priestle, J.P, Di Marco, S. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the complex of leech-derived tryptase inhibitor (LDTI) with trypsin and modeling of the LDTI-tryptase system.
Structure, 5, 1997
|
|
4A5T
 
 | | STRUCTURAL BASIS FOR THE CONFORMATIONAL MODULATION | | Descriptor: | CHLORIDE ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, PLASMINOGEN | | Authors: | Xue, Y, Bodin, C, Olsson, K. | | Deposit date: | 2011-10-28 | | Release date: | 2012-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structure of the Native Plasminogen Reveals an Activation-Resistant Compact Conformation.
J. Thromb. Haemost., 10, 2012
|
|
167D
 
 | |
1KLT
 
 | |
4AWN
 
 | | Structure of recombinant human DNase I (rhDNaseI) in complex with Magnesium and Phosphate. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parsiegla, G, Noguere, C, Santell, L, Lazarus, R.A, Bourne, Y. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Human DNase I Bound to Magnesium and Phosphate Ions Points to a Catalytic Mechanism Common to Members of the DNase I-Like Superfamily.
Biochemistry, 51, 2012
|
|
1AM5
 
 | |
2V9Y
 
 | | Human aminoimidazole ribonucleotide synthetase | | Descriptor: | PHOSPHORIBOSYLFORMYLGLYCINAMIDINE CYCLO-LIGASE, SULFATE ION | | Authors: | Welin, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Herman, M.D, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies of Tri-Functional Human Gart.
Nucleic Acids Res., 38, 2010
|
|
3QFR
 
 | | Crystal Structure of Human NADPH-Cytochrome P450 Reductase (R457H Mutant) | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S.P, Masters, B.S, Kim, J.-J.P. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QFS
 
 | | Crystal Structure of NADPH-Cytochrome P450 Reductase (FAD/NADPH domain) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH--cytochrome P450 reductase | | Authors: | Xia, C, Marohnic, C, Panda, S.P, Masters, B.S, Kim, J.-J.P. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1KR5
 
 | | Crystal structure of human L-isoaspartyl methyltransferase | | Descriptor: | Protein-L-isoaspartate O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ryttersgaard, C, Griffith, S.C, Sawaya, M.R, MacLaren, D.C, Clarke, S, Yeates, T.O. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human L-isoaspartyl methyltransferase.
J.Biol.Chem., 277, 2002
|
|
1ANN
 
 | | ANNEXIN IV | | Descriptor: | ANNEXIN IV, CALCIUM ION | | Authors: | Sutton, R.B, Sprang, S.R. | | Deposit date: | 1995-09-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three Dimensional Structure of Annexin IV
Annexins: Molecular Structure to Cellular Function, 1996
|
|
4ABJ
 
 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (IcA)SFTI-1(1,14), that was 1,5-disubstituted with 1,2,3- trizol to mimic a cis amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4ABI
 
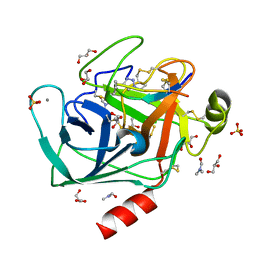 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (PtA)SFTI-1(1,14), that was 1,4-disubstituted with a 1,2,3- trizol to mimic a trans amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3QNF
 
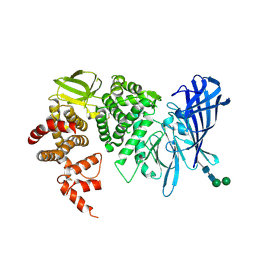 | | Crystal structure of the open state of human endoplasmic reticulum aminopeptidase 1 ERAP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, ZINC ION, ... | | Authors: | Vollmar, M, Kochan, G, Krojer, T, Harvey, D, Chaikuad, A, Allerston, C, Muniz, J.R.C, Raynor, J, Ugochukwu, E, Berridge, G, Wordsworth, B.P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QOA
 
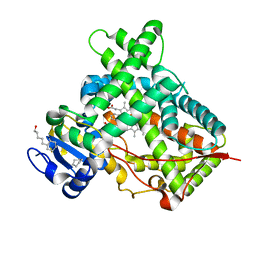 | | Crystal structure of a human cytochrome P450 2B6 (Y226H/K262R) in complex with the inhibitor 4-Benzylpyridine. | | Descriptor: | 4-benzylpyridine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-02-09 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cytochrome P450 2B6 Bound to 4-Benzylpyridine and 4-(4-Nitrobenzyl)pyridine: Insight into Inhibitor Binding and Rearrangement of Active Site Side Chains.
Mol.Pharmacol., 80, 2011
|
|
1KPR
 
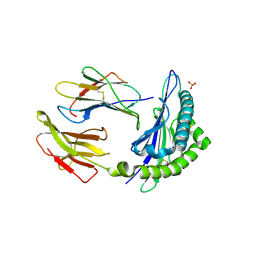 | | The human non-classical major histocompatibility complex molecule HLA-E | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ALPHA CHAIN, ... | | Authors: | Holmes, M.A, Strong, R.K. | | Deposit date: | 2002-01-02 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | HLA-E allelic variants: Correlating differential expression, peptide affinities, crystal structures and thermal stabilities
J.Biol.Chem., 278, 2003
|
|
4A7D
 
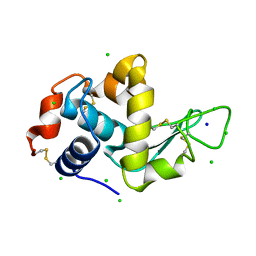 | | X-ray crystal structure of HEWL flash-cooled at high pressure | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Burkhardt, A, Warmer, M, Panneerselvam, S, Wagner, A, Reimer, R, Hohenberg, H, Meents, A. | | Deposit date: | 2011-11-14 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Fast High-Pressure Freezing of Protein Crystals in Their Mother Liquor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4ABF
 
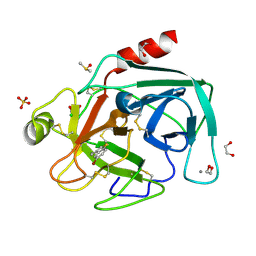 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-BROMO-1-BENZOTHIOPHEN-3-YL)METHANAMINE, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
1BNB
 
 | |
1I4A
 
 | | CRYSTAL STRUCTURE OF PHOSPHORYLATION-MIMICKING MUTANT T6D OF ANNEXIN IV | | Descriptor: | ANNEXIN IV, CALCIUM ION, SULFATE ION | | Authors: | Kaetzel, M.A, Mo, Y.D, Mealy, T.R, Campos, B, Bergsma-Schutter, W, Brisson, A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2001-02-20 | | Release date: | 2001-04-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation mutants elucidate the mechanism of annexin IV-mediated membrane aggregation.
Biochemistry, 40, 2001
|
|
4AEI
 
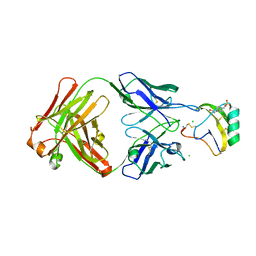 | | Crystal structure of the AaHII-Fab4C1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALPHA-MAMMAL TOXIN AAH2, CHLORIDE ION, ... | | Authors: | Fabrichny, I.P, Mondielli, G, Conrod, S, Martin-Eauclaire, M.F, Bourne, Y, Marchot, P. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into Antibody Sequestering and Neutralizing of Na+-Channel & [Alpha]-Type Modulator from Old-World Scorpion Venom
J.Biol.Chem., 287, 2012
|
|
1C2O
 
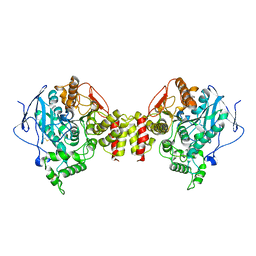 | | ELECTROPHORUS ELECTRICUS ACETYLCHOLINESTERASE | | Descriptor: | ACETYLCHOLINESTERASE | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Conformational flexibility of the acetylcholinesterase tetramer suggested by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
3QVV
 
 | |
3QXH
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ADP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Minor, C, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
