4LY9
 
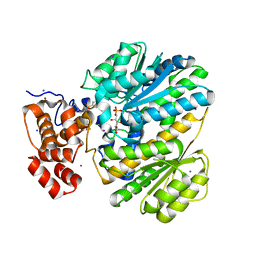 | | Human GKRP complexed to AMG-1694 [(2R)-1,1,1-trifluoro-2-{4-[(2S)-2-{[(3S)-3-methylmorpholin-4-yl]methyl}-4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol] and sorbitol-6-phosphate | | Descriptor: | (2R)-1,1,1-trifluoro-2-{4-[(2S)-2-{[(3S)-3-methylmorpholin-4-yl]methyl}-4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R. | | Deposit date: | 2013-07-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antidiabetic effects of glucokinase regulatory protein small-molecule disruptors.
Nature, 504, 2013
|
|
6SK8
 
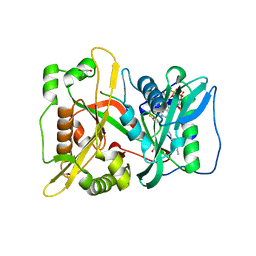 | | DeltaC3 C-terminal truncation of HsNMT1 in complex with MyrCoA and GDCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
7W3H
 
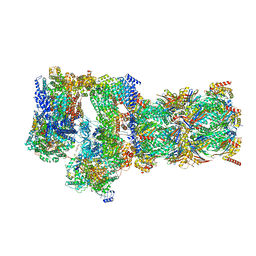 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
3OLY
 
 | |
8TS4
 
 | | Crystal structure of T. Brucei hypoxanthine guanine phosphoribosyltransferase in complex with [2S,4S]-4-Guanin-9-yl-2-(2-phosphonoethoxymethyl)-1-N-(3-phosphonopropionyl)pyrrolidine | | Descriptor: | (3-{(2S,4S)-4-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-[(2-phosphonoethoxy)methyl]pyrrolidin-1-yl}-3-oxopropyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Guddat, L.W, Guddat, L.W. | | Deposit date: | 2023-08-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Prolinol Containing Inhibitors of Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase: Rational Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
6SMF
 
 | |
7W39
 
 | | Structure of USP14-bound human 26S proteasome in state EA2.1_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
8TVN
 
 | | IRAK4 in complex with compound 23 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-[(1R,2S)-2-fluorocyclopropyl]-2-oxo-1,2-dihydropyridin-3-yl}-2-[(1R,4r)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-6-[(propan-2-yl)oxy]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Tiny Pocket Packs a Punch: Leveraging Pyridones for the Discovery of CNS-Penetrant Aza-indazole IRAK4 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
6ETY
 
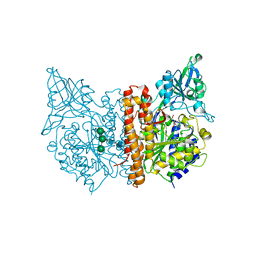 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor JHU3371 | | Descriptor: | (2~{S})-2-[[(2~{R})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]carbamoyloxy]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-10-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
8TU3
 
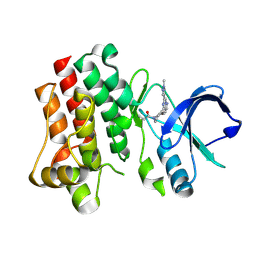 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 10 | | Descriptor: | 1-[(4R)-4-{[(6P,8S)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}azepan-1-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
8TVM
 
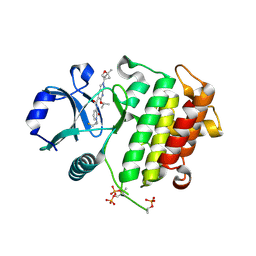 | | IRAK4 in complex with compound 24 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-[(1R,2R)-2-fluorocyclopropyl]-2-oxo-1,2-dihydropyridin-3-yl}-2-[(1R,4r)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-6-[(propan-2-yl)oxy]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Tiny Pocket Packs a Punch: Leveraging Pyridones for the Discovery of CNS-Penetrant Aza-indazole IRAK4 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
6VCT
 
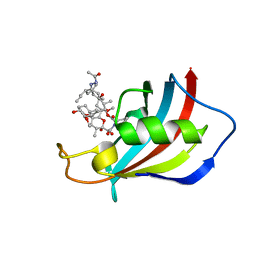 | | Mucor circinelloides FKBP12 protein bound with APX879 in C2221 space group | | Descriptor: | N'-[(3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,26aS)-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-1,20,21-trioxo-8-(prop-2-en-1-yl)-1,3,4,5,6,8,11,12,13,14,15,16,17,18,19,20,21,23,24,25,26,26a-docosahydro-7H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosin-7-ylidene]acetohydrazide, Peptidylprolyl isomerase | | Authors: | Gobeil, S, Spicer, L. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.
Mbio, 12, 2021
|
|
4MB5
 
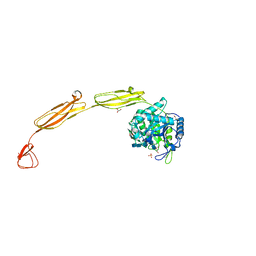 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5HQA
 
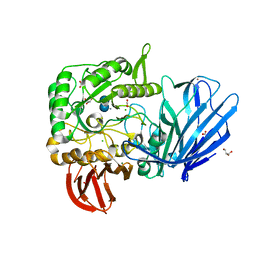 | | A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, CALCIUM ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
3P0G
 
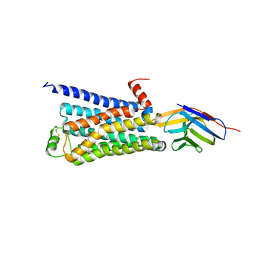 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
6CHL
 
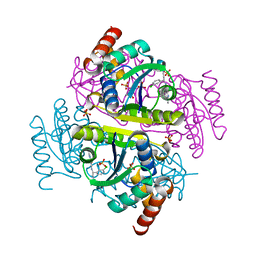 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with (R)-3-(3-chlorophenyl)-3-((5-methyl-7-oxo-4,7-dihydro-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)propanenitrile | | Descriptor: | (3R)-3-(3-chlorophenyl)-3-[(5-methyl-7-oxo-6,7-dihydro[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino]propanenitrile, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
6CHQ
 
 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with 2-benzyl-N-(3-chloro-4-methylphenyl)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | 2-benzyl-7-[(3-chloro-4-methylphenyl)amino]-5-methyl-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
3P5O
 
 | | Crystal Structure of the First Bromodomain of Human Brd4 in complex with IBET inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-N-ethylacetamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2010-10-09 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Suppression of inflammation by a synthetic histone mimic
Nature, 468, 2010
|
|
5H9P
 
 | | Crystal Structure of Human Galectin-3 CRD in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
7ZO3
 
 | | L1 metallo-beta-lactamase in complex with hydrolysed tebipenem | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[1-(4,5-dihydro-1,3-thiazol-2-yl)azetidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO4
 
 | | L1 metallo-beta-lactamase in complex with hydrolysed panipenem | | Descriptor: | (2R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S)-1-ethanimidoylpyrrolidin-3-yl]sulfanyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SODIUM ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
6J4W
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
8TRQ
 
 | | T cell recognition of citrullinated vimentin peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A07 TCR alpha chain, A07 TCR beta chain, ... | | Authors: | Loh, T.J, Lim, J.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
8THF
 
 | | SARS-CoV-2 BA.1 S-6P-no-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-07-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Universal subunit vaccine protects against multiple SARS-CoV-2 variants and SARS-CoV.
Npj Vaccines, 9, 2024
|
|
8A6E
 
 | | 100 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
