9UFR
 
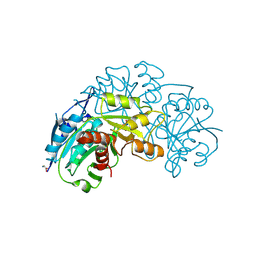 | | Crystal structure of L-asparaginase from Thermococcus Sibiricus double mutation D54G/T56Q | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, GLYCINE, ... | | Authors: | Matyuta, I.O, Varfolomeeva, L.A, Minyaev, M.E, Dumina, M, Zhdanov, D, Zhgun, A, Pokrovskaya, M, Aleksandrova, S, El'darov, M, Popov, V.O, Boyko, K.M. | | Deposit date: | 2025-04-10 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hyperthermophilic L-Asparaginase from Thermococcus sibiricus and Its Double Mutant with Increased Activity: Insights into Substrate Specificity and Structure.
Int J Mol Sci, 26, 2025
|
|
8GZ6
 
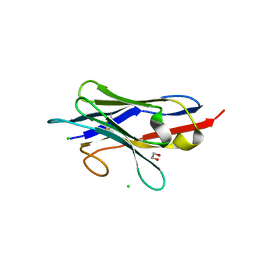 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nanobody P17 | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
6V7I
 
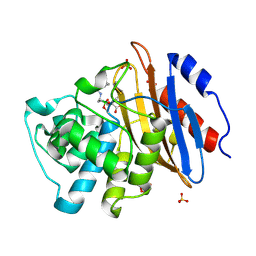 | | Structure of KPC-2 bound to Vaborbactam at 1.25 A | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, SULFATE ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-12-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Binding Kinetics of Vaborbactam in Class A beta-Lactamase Inhibition.
Antimicrob.Agents Chemother., 64, 2020
|
|
5IE1
 
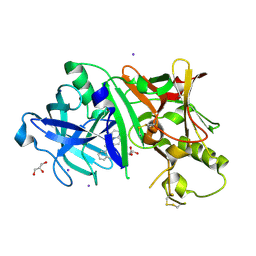 | | Crystal structure of BACE1 in complex with 3-(2-amino-6-(o-tolyl)quinolin-3-yl)-N-(3,3-dimethylbutyl)propanamide | | Descriptor: | 3-[2-amino-6-(2-methylphenyl)quinolin-3-yl]-N-(3,3-dimethylbutyl)propanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Jordan, J.B, Whittington, D.A, Bartberger, M.D, Sickmier, E.A, Chen, K, Cheng, Y, Judd, T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Fragment-Linking Approach Using (19)F NMR Spectroscopy To Obtain Highly Potent and Selective Inhibitors of beta-Secretase.
J.Med.Chem., 59, 2016
|
|
9V4P
 
 | | Cryo-EM structure of A. thaliana MET1(610-1534) bound covalently to a hemi-mCpG DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*GP*GP*AP*TP*(C49)P*GP*TP*AP*TP*GP*TP*CP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*A*TP*GP*AP*CP*AP*TP*AP*(5CM)P*GP*AP*TP*CP*CP*AP*AP*TP*C)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Zhang, Z, Li, W, Liu, Y, Du, J. | | Deposit date: | 2025-05-24 | | Release date: | 2025-11-26 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Structural insights into plant DNA CG methylation maintenance by MET1.
Plant Cell, 37, 2025
|
|
7YRG
 
 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
9OJX
 
 | | Crystal structure of E. coli ApaH in complex with GDP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bis(5'-nucleosyl)-tetraphosphatase [symmetrical], GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2025-05-08 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | ApaH decaps Np 4 N-capped RNAs in two alternative orientations.
Nat.Chem.Biol., 2025
|
|
9SEA
 
 | |
9OW2
 
 | | Crystal Structure of the Surface Protein (CD630_07380) from Clostridium difficile Strain 630 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-06-02 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Surface Protein (CD630_07380) from Clostridium difficile Strain 630.
To Be Published
|
|
9EW1
 
 | | Ternary structure of 14-3-3s, CRAF phosphopeptide (pS259) and compound 79 (1124379). | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-1-[8-(4-iodophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]ethanone, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
6P5P
 
 | | Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Anti-Tumor Agent | | Descriptor: | 2-[(2S)-1-azabicyclo[2.2.2]octan-2-yl]-6-(5-methyl-1H-pyrazol-4-yl)thieno[3,2-d]pyrimidin-4(3H)-one, Rho-associated protein kinase 2 | | Authors: | Hoffman, I.D, Skene, R.J. | | Deposit date: | 2019-05-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 Inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Antitumor Agent.
J.Med.Chem., 63, 2020
|
|
5IFS
 
 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
9R5Z
 
 | | Crystal structure of JAK3 with GCL258 | | Descriptor: | 1-phenylurea, 3-(3-cyclohexyl-3,8,10-triazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-5-yl)benzenesulfonyl fluoride, Tyrosine-protein kinase JAK3 | | Authors: | Wang, G.Q, Chaikuad, A, Hillebrand, L, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-05-11 | | Release date: | 2025-08-20 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A twist in the tale: shifting from covalent targeting of a tyrosine in JAK3 to a lysine in MK2.
Rsc Med Chem, 16, 2025
|
|
7T5H
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, wild type | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphoprotein, ... | | Authors: | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
7PJG
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2R,4R)-N-[[4-(4-cyclopropylbuta-1,3-diynyl)phenyl]methyl]-1-(2-methylpropanoyl)-4-[[(2S,4R)-4-oxidanylpyrrolidin-2-yl]carbonylamino]pyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
9V8D
 
 | | PPARgamma ligand-binding domain in complex with PG08 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PG08, ... | | Authors: | Sigal, M, Okada, C, Katoh, T, Suga, H, Sengoku, T. | | Deposit date: | 2025-05-29 | | Release date: | 2025-11-05 | | Last modified: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | De Novo Discovery of alpha , alpha-Disubstituted alpha-amino Acid-Containing alpha-helical Peptides as Competitive PPAR gamma PPI Inhibitors.
J.Am.Chem.Soc., 147, 2025
|
|
9PLN
 
 | | Locally-refined structure of alpha2a adrenergic receptor in complex with Go heterotrimer, scFv16, and N-(5-methylnaphthalen-1-yl)pyridin-4-amine (compound 4905) | | Descriptor: | Alpha-2A adrenergic receptor, N-(5-methylnaphthalen-1-yl)pyridin-4-amine | | Authors: | Srinivasan, K, Xu, X, Mailhot, O, Manglik, A, Shoichet, B. | | Deposit date: | 2025-07-15 | | Release date: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Toward a Random Background for Ligand Optimization
To Be Published
|
|
9OCE
 
 | | 2.48A cryo-EM structure of the Measles Virus L-P-C in complex with ERdRp-0519 | | Descriptor: | 2-methyl-~{N}-[4-[(2~{S})-2-(2-morpholin-4-ylethyl)piperidin-1-yl]sulfonylphenyl]-5-(trifluoromethyl)pyrazole-3-carboxamide, Phosphoprotein, Protein C, ... | | Authors: | Liu, B, Wang, D, Yang, G. | | Deposit date: | 2025-04-24 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis of measles virus polymerase inhibition by nonnucleoside inhibitor ERDRP-0519.
Nat Commun, 16, 2025
|
|
9BCY
 
 | | Crystal structure of Mayaro virus capsid C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Structural polyprotein | | Authors: | Bezerra, E.H.S, Scorsato, V, Tonoli, C.C, Benedetti, C.E, Marques, R.E. | | Deposit date: | 2024-04-10 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of Mayaro virus capsid C-terminal domain
To Be Published
|
|
7PK8
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2S,4S)-4-fluoranyl-N-[(3R,5R)-5-[[[4-[2-(4-methylphenyl)ethynyl]phenyl]carbonylamino]methyl]-1-(2-methylpropanoyl)pyrrolidin-3-yl]pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
7ZT2
 
 | | Structure of E8 TCR in complex with human MR1 bound to 5-OP-RU | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, ... | | Authors: | Karuppiah, V, Srikannathasan, V, Robinson, R.A. | | Deposit date: | 2022-05-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Promiscuous recognition of MR1 drives self-reactive mucosal-associated invariant T cell responses.
J.Exp.Med., 220, 2023
|
|
3DAS
 
 | | Structure of the PQQ-bound form of Aldose Sugar Dehydrogenase (Adh) from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Southall, S.M, Doel, J.J, Oubrie, A, Richardson, D.J. | | Deposit date: | 2008-05-30 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Enzymatic Characterization of a Thermostable PQQ-dependent Soluble Aldose Sugar Dehydrogenase
To be Published
|
|
9OIN
 
 | | The von Hippel Lindau-ElonginB-ElonginC (VCB) complex with fragment 13 | | Descriptor: | 1-(propan-2-yl)-4-[(pyridin-2-yl)methyl]piperazine, Elongin-B, Elongin-C, ... | | Authors: | Amporndanai, K, Katinas, J.M, Chopra, A, Fesik, S.W. | | Deposit date: | 2025-05-06 | | Release date: | 2025-07-09 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | NMR-Based Fragment Screen of the von Hippel-Lindau Elongin C&B Complex.
Acs Med.Chem.Lett., 16, 2025
|
|
4XNL
 
 | | X-ray structure of AlgE2 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Ma, P, Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9NXJ
 
 | | The GH43 domain of an alpha-l-arabinofuranosidase (AtAbf43C_GH43) from Acetivibrio thermocellus DSM1313 | | Descriptor: | Glycoside hydrolase family 43, SULFATE ION, alpha-L-arabinofuranose | | Authors: | Galindo, J.L, Jeffrey, P.D, Conway, J.M. | | Deposit date: | 2025-03-25 | | Release date: | 2025-08-20 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Functional and structural characterization of AtAbf43C: an exo-1,5-alpha-L-arabinofuranosidase from Acetivibrio thermocellus DSM1313.
Biochem.J., 482, 2025
|
|
