7ZLW
 
 | | NME1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
8DCF
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Cu2+ and GTP | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DCA
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Co2+ and GTP | | Descriptor: | COBALT (II) ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
6Z5D
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004082 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 11-methyl-8,11,14,18,19,22-hexazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2(23),3,5,15(22),16,19-heptaen-7-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004082
To Be Published
|
|
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2022-02-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8D80
 
 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
9Q7W
 
 | |
7ZJO
 
 | |
9C7Y
 
 | | Structure Of Respiratory Syncytial Virus Polymerase in complex with JNJ-2729 | | Descriptor: | (2S)-1,1,1-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-3-[(4M)-4-(8-methoxyquinolin-6-yl)-1H-1,2,3-triazol-1-yl]propan-2-ol, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yin, Y, Tran, M.T, Yu, X, Jonckers, T. | | Deposit date: | 2024-06-11 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure-Activity Relationship of Oxacyclo- and Triazolo-Containing Respiratory Syncytial Virus Polymerase Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
5K33
 
 | | Crystal structure of extracellular domain of HER2 in complex with Fcab STAB19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ig gamma-1 chain C region, ... | | Authors: | Humm, A, Lobner, E, Goritzer, K, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
9SA2
 
 | | The RING domain of IDOL | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, ZINC ION | | Authors: | Bradshaw, W.J, He, L, Guenther, F, Murphy, E.J. | | Deposit date: | 2025-08-07 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The RING domain of IDOL
To Be Published
|
|
8AN8
 
 | | Crystal structure of wild-type c-MET bound by compound 7. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
6ZA3
 
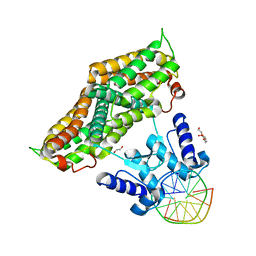 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (C2221 space group) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
9CED
 
 | | SARS-CoV-2 3CL Protease complexed with covalent inhibitor VK13 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-{[(2S)-1-hydroxy-3-(2-oxo-1,2-dihydropyridin-3-yl)propan-2-yl]amino}-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Hoffpauir, Z.A, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2024-06-26 | | Release date: | 2025-01-15 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Dual Inhibitors of SARS-CoV-2 3CL Protease and Human Cathepsin L Containing Glutamine Isosteres Are Anti-CoV-2 Agents.
J.Am.Chem.Soc., 147, 2025
|
|
9CF9
 
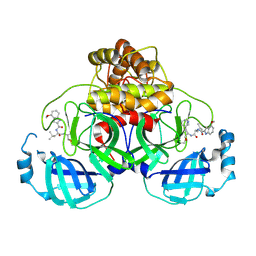 | | SARS-CoV-2 3CL Protease complexed with covalent inhibitor BC787 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-{[(2S)-1-hydroxy-3-(pyridin-3-yl)propan-2-yl]amino}-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Hoffpauir, Z.A, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2024-06-27 | | Release date: | 2025-01-15 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual Inhibitors of SARS-CoV-2 3CL Protease and Human Cathepsin L Containing Glutamine Isosteres Are Anti-CoV-2 Agents.
J.Am.Chem.Soc., 147, 2025
|
|
8C6T
 
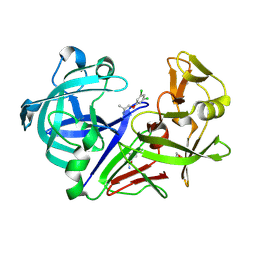 | | Fragment screening hit IV bound to endothiapepsin | | Descriptor: | 1-[3,5-bis(chloranyl)phenoxy]propan-2-amine, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
9CFB
 
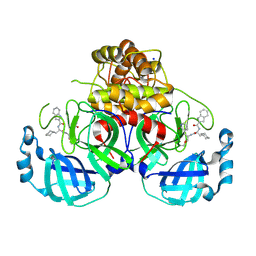 | | SARS-CoV-2 3CL Protease complexed with covalent inhibitor BC674 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-{[(2S)-1-hydroxy-3-(2-oxo-1,2-dihydropyridin-3-yl)propan-2-yl]amino}-1-oxopropan-2-yl]-1H-indole-2-carboxamide, SODIUM ION | | Authors: | Hoffpauir, Z.A, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2024-06-27 | | Release date: | 2025-01-15 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual Inhibitors of SARS-CoV-2 3CL Protease and Human Cathepsin L Containing Glutamine Isosteres Are Anti-CoV-2 Agents.
J.Am.Chem.Soc., 147, 2025
|
|
7Q2I
 
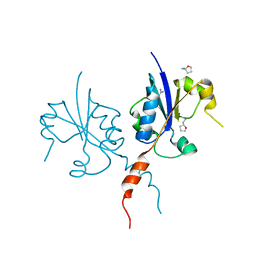 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Tetrahydrofurfurylamine | | Descriptor: | 1-[(2R)-oxolan-2-yl]methanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
6ZEL
 
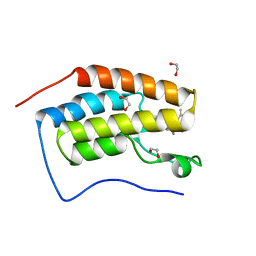 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F5 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-dimethyl-4-[(6-methylpyrimidin-4-yl)sulfanylmethyl]-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F5
To Be Published
|
|
8H8Q
 
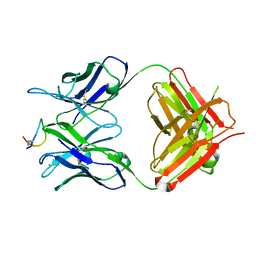 | | Fab-amyloid beta fragment complex at neutral pH | | Descriptor: | CHLORIDE ION, Fab, GLN-LYS-CYS-VAL-PHE-PHE-ALA-GLU-ASP-VAL-GLY-SER-ASN-CYS-GLY, ... | | Authors: | Kita, A, Irie, K, Irie, Y, Matsushima, Y, Miki, K. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the binding site and immunoreactivity of anti-A beta antibody 11A1: Comparison with the toxic conformation-specific TxCo-1 antibody.
Biochem.Biophys.Res.Commun., 758, 2025
|
|
8F4B
 
 | |
6ZGL
 
 | |
8CJD
 
 | | AetF, a single-component flavin-dependent tryptophan halogenase | | Descriptor: | 1,2-ETHANEDIOL, AetF, CALCIUM ION, ... | | Authors: | Gafe, S, Niemann, H.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of regioselective tryptophan dibromination by the single-component flavin-dependent halogenase AetF.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8C2G
 
 | |
8HAR
 
 | | SAH-bound C-Methyltransferase Fur6 from Streptomyces sp. KO-3988 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fur6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Noguchi, T, Nagata, R, Tomita, T, Kuzuyama, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-01 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Biosynthesis of the tetrahydroxynaphthalene-derived meroterpenoid furaquinocin via reductive deamination and intramolecular hydroalkoxylation of an alkene.
Chem Sci, 2025
|
|
