1ZT9
 
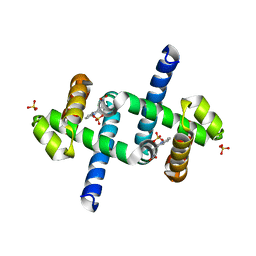 | | E. coli trp repressor, tetragonal crystal form | | Descriptor: | SULFATE ION, TRYPTOPHAN, Trp operon repressor | | Authors: | Lawson, C.L, Chin, A.S, Benoff, B, Yung, B.H. | | Deposit date: | 2005-05-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two association modes for E. coli trp repressor dimer-dimer interactions
To be Published
|
|
8SUB
 
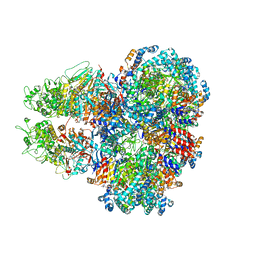 | | E. coli SIR2-HerA complex (dodecamer SIR2 pentamer HerA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside triphosphate hydrolase, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-05-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8SUW
 
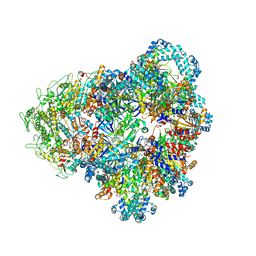 | | E. coli SIR2-HerA complex (dodecamer SIR2 bound 4 protomers of HerA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside triphosphate hydrolase, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-05-13 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8UAE
 
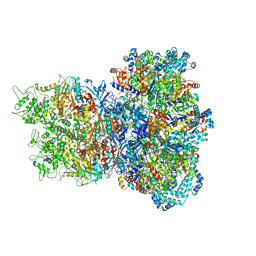 | | E. coli Sir2_HerA complex (12:6) with ATPgamaS | | Descriptor: | MAGNESIUM ION, Nucleoside triphosphate hydrolase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8TXO
 
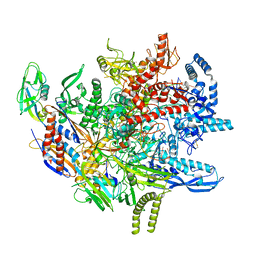 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
8VRG
 
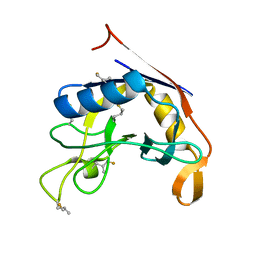 | | E. coli peptidyl-prolyl cis-trans isomerase containing delta1-monofluoro-leucines | | Descriptor: | Peptidyl-prolyl cis-trans isomerase B | | Authors: | Frkic, R.L, Jackson, C.J. | | Deposit date: | 2024-01-22 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Conformational Preferences of the Non-Canonical Amino Acids (2 S ,4 S )-5-Fluoroleucine, (2 S ,4 R )-5-Fluoroleucine, and 5,5'-Difluoroleucine in a Protein.
Biochemistry, 63, 2024
|
|
6G8B
 
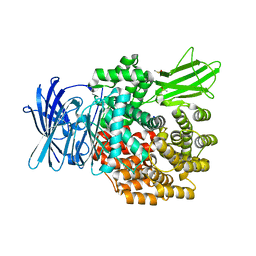 | | E. coli Aminopeptidase N solved by Native SAD from a dataset collected in 60 second with JUNGFRAU detector | | Descriptor: | Aminopeptidase N, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Leonarski, F, Olieric, V, Redford, S, Wang, M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Fast and accurate data collection for macromolecular crystallography using the JUNGFRAU detector.
Nat. Methods, 15, 2018
|
|
3KOY
 
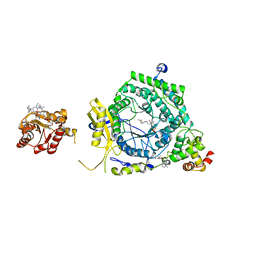 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Aerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
8EDQ
 
 | | E. coli pyruvate kinase (PykF) I264F | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyruvate kinase, SULFATE ION | | Authors: | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
2Q96
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A18 | | Descriptor: | 5-(2-CHLOROBENZYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
3KOZ
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Anaerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
2Q94
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A04 | | Descriptor: | 5-[2-(TRIFLUOROMETHOXY)PHENYL]-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q93
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor B21 | | Descriptor: | 5-(2-METHOXYPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q95
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A05 | | Descriptor: | 5-(2-CHLORO-4-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q92
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor B23 | | Descriptor: | 5-(2-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
1HN1
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION, SODIUM ION | | Authors: | Juers, D.H, Matthews, B.W. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversible lattice repacking illustrates the temperature dependence of macromolecular interactions.
J.Mol.Biol., 311, 2001
|
|
5KNS
 
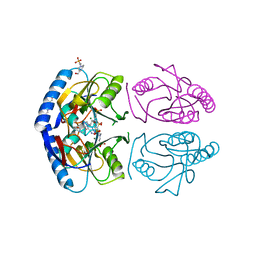 | | E coli hypoxanthine guanine phosphoribosyltransferase in complexed with 9-[(N-phosphonoethyl-N-phosphonoethoxyethyl)-2-aminoethyl]hypoxanthine | | Descriptor: | (2-{[2-(6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl](2-{[(E)-2-phosphonoethenyl]oxy}ethyl)amino}ethyl)phosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
6B88
 
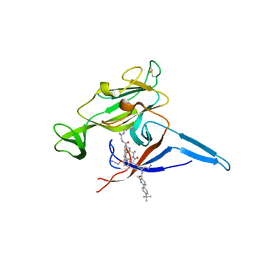 | | E. coli LepB in complex with GNE0775 ((4S,7S,10S)-10-((S)-4-amino-2-(2-(4-(tert-butyl)phenyl)-4-methylpyrimidine-5-carboxamido)-N-methylbutanamido)-16,26-bis(2-aminoethoxy)-N-(2-iminoethyl)-7-methyl-6,9-dioxo-5,8-diaza-1,2(1,3)-dibenzenacyclodecaphane-4-carboxamide) | | Descriptor: | (8S,11S,14S)-14-{[(2S)-4-amino-2-{[2-(4-tert-butylphenyl)-4-methylpyrimidine-5-carbonyl]amino}butanoyl](methyl)amino}-3,18-bis(2-aminoethoxy)-N-[(2Z)-2-iminoethyl]-11-methyl-10,13-dioxo-9,12-diazatricyclo[13.3.1.1~2,6~]icosa-1(19),2(20),3,5,15,17-hexaene-8-carboxamide, PENTAETHYLENE GLYCOL, Signal peptidase I | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Optimized arylomycins are a new class of Gram-negative antibiotics.
Nature, 561, 2018
|
|
7R1W
 
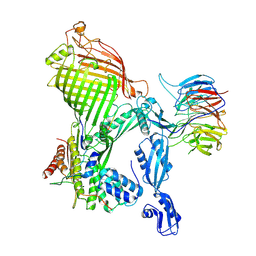 | | E. coli BAM complex (BamABCDE) bound to dynobactin A | | Descriptor: | Dynobactin A, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Hiller, S, Maier, T. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
7MR3
 
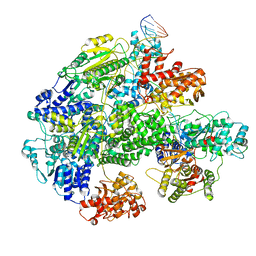 | | Cryo-EM structure of RecBCD-DNA complex with docked RecBNuc and stabilized RecD | | Descriptor: | DNA (60-MER), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Hao, L, Zhang, R, Lohman, T.M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.
J.Mol.Biol., 433, 2021
|
|
7MR4
 
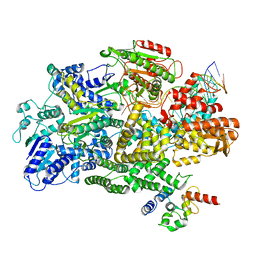 | | Cryo-EM structure of RecBCD-DNA complex with undocked RecBNuc and flexible RecD | | Descriptor: | DNA (60-MER), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Hao, L, Zhang, R, Lohman, T.M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.
J.Mol.Biol., 433, 2021
|
|
3VIC
 
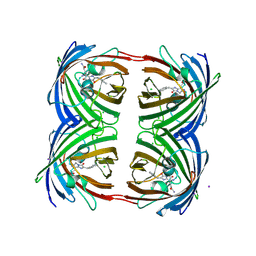 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Byres, E, Wilce, M, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
1JZ6
 
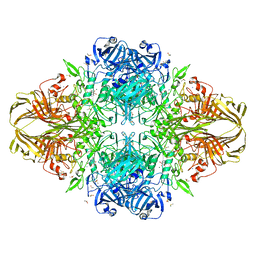 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTO-TETRAZOLE | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Juers, D.H, Heightman, T.D, Vasella, A, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
1JZ5
 
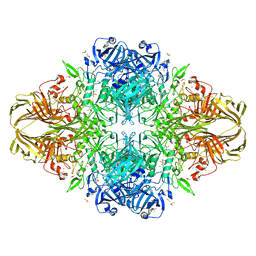 | |
1JYW
 
 | |
