1VTC
 
 | |
1VTI
 
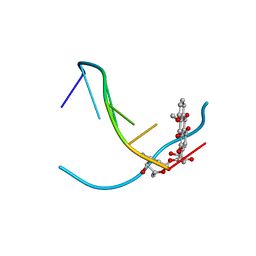 | | DNA-DRUG INTERACTIONS: THE CRYSTAL STRUCTURES OF D(TGATCA) COMPLEXED WITH DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3') | | Authors: | Nunn, C.M, Van Meervelt, L, Zhang, S, Moore, M.H, Kennard, O. | | Deposit date: | 1992-03-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DNA-Drug Interactions: The Crystal Structures of d(TGTACA) and d(TGATCA) Complexed with Daunomycin
J.Mol.Biol., 222, 1991
|
|
1VYN
 
 | |
1VTW
 
 | | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of D(M(5)CGTAM(5)CG) | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*TP*AP*(CH3)CP*G)-3') | | Authors: | Wang, A.H.-J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of d(m(5)CGTAm(5)CG)
Cell(Cambridge,Mass.), 37, 1984
|
|
2E67
 
 | |
1VZY
 
 | | Crystal structure of the Bacillus subtilis HSP33 | | Descriptor: | 33 KDA CHAPERONIN, ACETATE ION, ZINC ION | | Authors: | Janda, I.K, Devedjiev, Y, Derewenda, U, Dauter, Z, Bielnicki, J, Cooper, D.R, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-29 | | Release date: | 2004-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism.
Structure, 12, 2004
|
|
1VDF
 
 | |
2E9Q
 
 | | Recombinant pro-11S globulin of pumpkin | | Descriptor: | 11S globulin subunit beta, CHLORIDE ION, PHOSPHATE ION | | Authors: | Fukuda, T, Prak, K, Itoh, T, Masuda, T, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
1VJO
 
 | |
1VK6
 
 | |
2EJ8
 
 | | Crystal structure of APPL1 PTB domain at 1.8A | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Tanabe, H, Hosaka, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of APPL1 PTB domain at 1.8A
To be Published
|
|
2EA4
 
 | | h-MetAP2 complexed with A797859 | | Descriptor: | 2-(2-AMINOETHOXY)-3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EEO
 
 | |
2F57
 
 | | Crystal Structure Of The Human P21-Activated Kinase 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE, Serine/threonine-protein kinase PAK 7 | | Authors: | Eswaran, J, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Bray, J, Das, S, Savitsky, P, Smee, C, Burgess, N, Fedorov, O, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs.
Structure, 15, 2007
|
|
2F91
 
 | | 1.2A resolution structure of a crayfish trypsin complexed with a peptide inhibitor, SGTI | | Descriptor: | CADMIUM ION, CHLORIDE ION, Serine protease inhibitor I/II, ... | | Authors: | Fodor, K, Harmat, V, Hetenyi, C, Kardos, J, Antal, J, Perczel, A, Patthy, A, Katona, G, Graf, L. | | Deposit date: | 2005-12-05 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Enzyme:Substrate Hydrogen Bond Shortening during the Acylation Phase of Serine Protease Catalysis.
Biochemistry, 45, 2006
|
|
2FFI
 
 | | Crystal Structure of Putative 2-Pyrone-4,6-Dicarboxylic Acid Hydrolase from Pseudomonas putida, Northeast Structural Genomics Target PpR23. | | Descriptor: | 2-pyrone-4,6-dicarboxylic acid hydrolase, putative, PHOSPHATE ION | | Authors: | Forouhar, F, Su, M, Jayaraman, S, Conover, K, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Putative 2-Pyrone-4,6-Dicarboxylic Acid Hydrolase from Pseudomonas putida, Northeast Structural Genomics Target PpR23.
To be Published
|
|
2FO7
 
 | |
2FV4
 
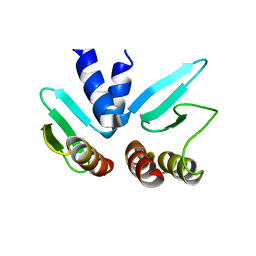 | |
2FPB
 
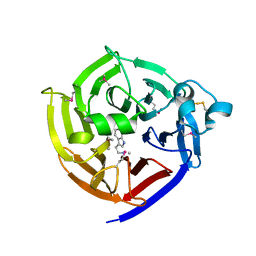 | |
2FSA
 
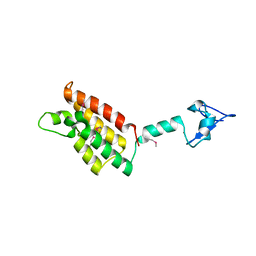 | |
2FSQ
 
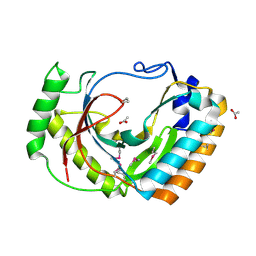 | | Crystal Structure of the Conserved Protein of Unknown Function ATU0111 from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETIC ACID, Atu0111 protein | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Zheng, H, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Atu0111 from Agrobacterium tumefaciens str. C58
To be Published
|
|
2G1D
 
 | | Solution Structure of Ribosomal Protein S24E from Thermoplasma acidophilum | | Descriptor: | 30S ribosomal protein S24e | | Authors: | Jeon, B.-Y, Hong, E.-M, Jung, J.-W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2006-02-14 | | Release date: | 2007-02-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of TA1092, a ribosomal protein S24e from Thermoplasma acidophilum
Proteins, 64, 2006
|
|
2G1A
 
 | | Crystal structure of the complex between Apha class B acid phosphatase/phosphotransferase | | Descriptor: | Class B acid phosphatase, MAGNESIUM ION, {[2-(6-AMINO-9H-PURIN-9-YL)ETHOXY]METHYL}PHOSPHONIC ACID | | Authors: | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | Deposit date: | 2006-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex between Apha class B acid phosphatase/phosphotransferase
To be published
|
|
2FP8
 
 | |
2FR2
 
 | |
