4YHM
 
 | | Reversal Agent for Dabigatran | | Descriptor: | N-[(2-{[(4-carbamimidoylphenyl)amino]methyl}-1-methyl-1H-benzimidazol-5-yl)carbonyl]-N-pyridin-2-yl-beta-alanine, aDabi-Fab2b heavy chain, aDabi-Fab2b light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
4YHT
 
 | | bRaf complexed with an inhibitor | | Descriptor: | 3-[(5-chloro-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-N-methyl-4-(morpholin-4-yl)benzenesulfonamide, GLYCEROL, Serine/threonine-protein kinase B-raf | | Authors: | Shewchuk, L.M, Lawhorn, B.G. | | Deposit date: | 2015-02-27 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | GSK114: A selective inhibitor for elucidating the biological role of TNNI3K.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5JIL
 
 | | Crystal structure of rat coronavirus strain New-Jersey Hemagglutinin-Esterase in complex with 4N-acetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8B99
 
 | | Crystal structure of JAK2 JH2-V617F in complex with JNJ-7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8INZ
 
 | | Cryo-EM structure of human HCN3 channel in apo state | | Descriptor: | 4-[[(2~{S},4~{a}~{R},6~{S},8~{a}~{S})-6-[(4~{S},5~{R})-4-[(2~{S})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structure of human HCN3 channel and its regulation by cAMP.
J.Biol.Chem., 300, 2024
|
|
6C1S
 
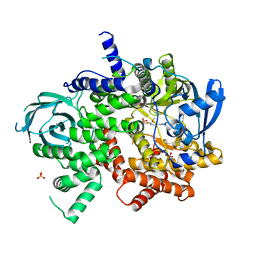 | | Phosphoinositide 3-Kinase gamma bound to an pyrrolopyridinone Inhibitor | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, {4-[2-(5,6-dimethoxypyridin-3-yl)-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-1H-pyrazol-1-yl}acetonitrile | | Authors: | Jacobs, M.D, Griffin, J.P. | | Deposit date: | 2018-01-05 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Synthesis of a Novel Series of Orally Bioavailable, CNS-Penetrant, Isoform Selective Phosphoinositide 3-Kinase gamma (PI3K gamma ) Inhibitors with Potential for the Treatment of Multiple Sclerosis (MS).
J. Med. Chem., 61, 2018
|
|
8BSG
 
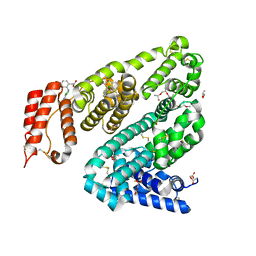 | | COMPLEX OF LEPORINE SERUM ALBUMIN WITH DICLOFENAC | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Bujacz, A, Talaj, J, Zielinski, K. | | Deposit date: | 2022-11-25 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Int J Mol Sci, 24, 2023
|
|
4L3Q
 
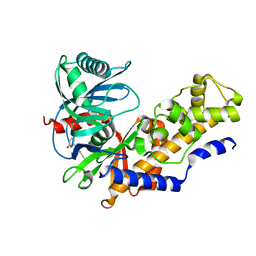 | | Crystal structure of glucokinase-activator complex | | Descriptor: | 6-{3-[(1-methyl-1H-imidazol-2-yl)sulfanyl]phenyl}pyridin-2(1H)-one, Glucokinase, alpha-D-glucopyranose | | Authors: | Gajiwala, K.S, Filipski, K.J. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrimidone-based series of glucokinase activators with alternative donor-acceptor motif.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6C96
 
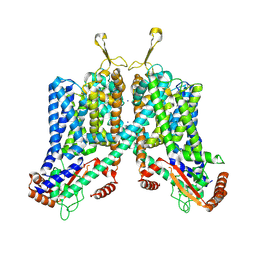 | | Cryo-EM structure of mouse TPC1 channel in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
7QOC
 
 | |
7QOE
 
 | |
6C53
 
 | | Cryo-EM structure of the Type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zheng, W, Wang, F, Luna-Rico, A, Francetic, O, Hultgren, S.J, Egelman, E.H. | | Deposit date: | 2018-01-13 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional role of the type 1 pilus rod structure in mediating host-pathogen interactions.
Elife, 7, 2018
|
|
7QOD
 
 | |
7JU8
 
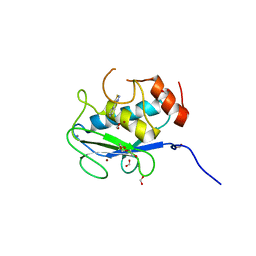 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
6S73
 
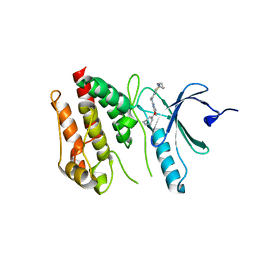 | | Crystal structure of Nek7 SRS mutant bound to compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Byrne, M.J, Bhatia, C, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
6YRA
 
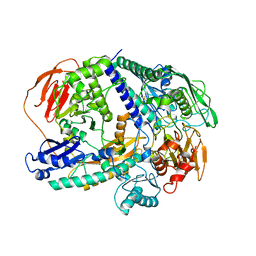 | |
8HZD
 
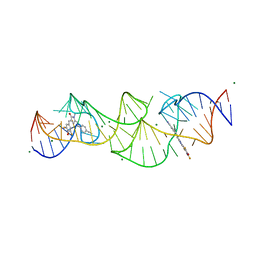 | | A new fluorescent RNA aptamer bound with N618 | | Descriptor: | 4-[(~{E})-2-[(4~{Z})-4-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-1-methyl-5-oxidanylidene-imidazol-2-yl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, RNA (36-MER) | | Authors: | Song, Q.Q, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
4YV3
 
 | |
6BL0
 
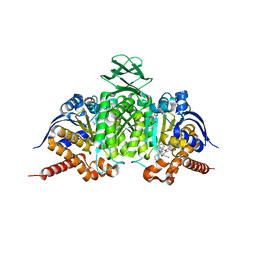 | | Novel Modes of Inhibition of Wild-Type IDH1:Direct Covalent Modification of His315 with Cmpd11 | | Descriptor: | (5aS,6S,8S,9aS)-2-(benzenecarbonyl)-6-methyl-7-oxo-9a-phenyl-4,5,5a,6,7,8,9,9a-octahydro-2H-benzo[g]indazole-8-carbonitrile, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
7BW6
 
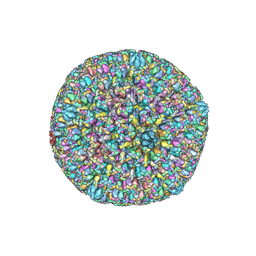 | | Varicella-zoster virus capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Wang, P.Y, Qi, J.X, Liu, C.C, Sun, J.Q. | | Deposit date: | 2020-04-13 | | Release date: | 2020-09-23 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the varicella-zoster virus A-capsid.
Nat Commun, 11, 2020
|
|
2O9I
 
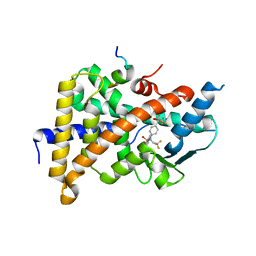 | | Crystal Structure of the Human Pregnane X Receptor LBD in complex with an SRC-1 coactivator peptide and T0901317 | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Nuclear Receptor Coactivator 1 isoform 3, Orphan nuclear receptor PXR | | Authors: | Xue, Y, Redinbo, M.R. | | Deposit date: | 2006-12-13 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism.
Bioorg.Med.Chem., 15, 2007
|
|
8JBR
 
 | | Structure of McyA2-CAPCP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, McyA protein | | Authors: | Peng, Y.J. | | Deposit date: | 2023-05-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modular catalytic activity of nonribosomal peptide synthetases depends on the dynamic interaction between adenylation and condensation domains.
Structure, 32, 2024
|
|
4YHI
 
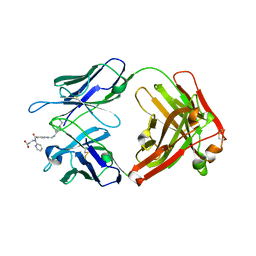 | | Reversal Agent for Dabigatran | | Descriptor: | N-[(2-{[(4-carbamimidoylphenyl)amino]methyl}-1-methyl-1H-benzimidazol-5-yl)carbonyl]-N-pyridin-2-yl-beta-alanine, aDabi-Fab2a heavy chain, aDabi-Fab2a light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
4YHN
 
 | | Dabigatran Reversal Agent | | Descriptor: | aDabi-Fab3 heavy chain, aDabi-Fab3 light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
7BR7
 
 | | Epstein-Barr virus, C1 portal-proximal penton vertex, CATC binding | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
