8FQZ
 
 | | Carbonic Anhydrase II in complex with the alkyl urea 3j | | Descriptor: | 4-hydroxy-3-{[(4-hydroxybutyl)carbamoyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FLP
 
 | | NMR Solution Structure of LvIC analogue | | Descriptor: | Alpha-conotoxin LvIC analogue | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Engineering of LvIC, an alpha 4/4-Conotoxin That Selectively Blocks Rat alpha 6/ alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 66, 2023
|
|
8Q0R
 
 | | X-ray structure of MNEI mutant Mut9 (E23A, C41A, Y65R, S76Y) | | Descriptor: | ACETATE ION, Monellin chain B,Monellin chain A, SULFATE ION | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
8FQY
 
 | | Carbonic Anhydrase II in complex with the alkyl urea 3h | | Descriptor: | 4-hydroxy-3-({[2-(pyridin-2-yl)ethyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FR4
 
 | |
8Q0S
 
 | | X-ray structure of the single chain monellin derivative MNEI | | Descriptor: | ACETATE ION, GLYCEROL, Monellin chain B,Monellin chain A, ... | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
8FR2
 
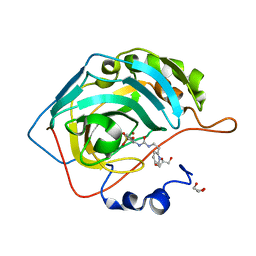 | |
6HNC
 
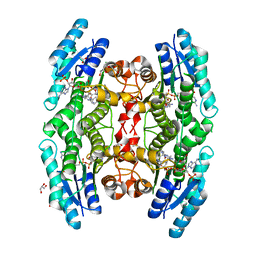 | | Trypanosoma brucei PTR1 in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-09-14 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Development of Cycloguanil Derivatives asTrypanosoma bruceiPteridine-Reductase-1 Inhibitors.
Acs Infect Dis., 5, 2019
|
|
6RHK
 
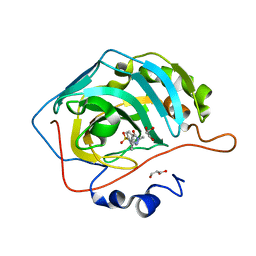 | | The crystal structure of human carbonic anhydrase II in complex with 4-(3-benzylimidazolidine-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-[3-(phenylmethyl)imidazolidin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
7QB1
 
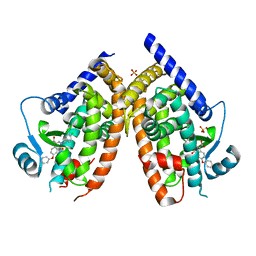 | | PPARg in complex with inhibitor | | Descriptor: | 3-[(3,4-dichlorophenyl)methyl]-4-oxidanylidene-6-phenoxy-phthalazine-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Petersen, J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery by Virtual Screening of an Inhibitor of CDK5-Mediated PPAR gamma Phosphorylation.
Acs Med.Chem.Lett., 13, 2022
|
|
6HZA
 
 | |
6HLB
 
 | |
6HLE
 
 | |
6RG0
 
 | | Structure of pdxj | | Descriptor: | Pyridoxine 5'-phosphate synthase | | Authors: | Rohweder, B, Rajendran, C, Sterner, R. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.074 Å) | | Cite: | Library Selection with a Randomized Repertoire of ( beta alpha )8-Barrel Enzymes Results in Unexpected Induction of Gene Expression.
Biochemistry, 58, 2019
|
|
8FU7
 
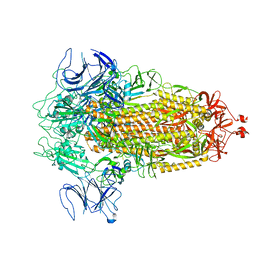 | | Structure of Covid Spike variant deltaN135 in fully closed form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
6HOW
 
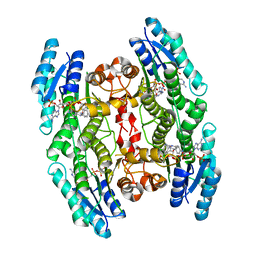 | | Trypanosoma brucei PTR1 in complex with the triazine inhibitor 2a (F219). | | Descriptor: | (2~{R})-1-(3,4-dichlorophenyl)-2-(4-nitrophenyl)-2~{H}-1,3,5-triazine-4,6-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Development of Cycloguanil Derivatives asTrypanosoma bruceiPteridine-Reductase-1 Inhibitors.
Acs Infect Dis., 5, 2019
|
|
8SWH
 
 | |
6HQG
 
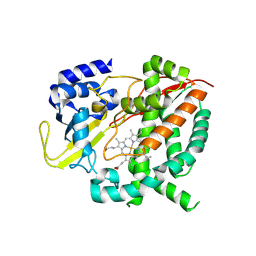 | | Cytochrome P450-153 from Phenylobacterium zucineum | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
8FU8
 
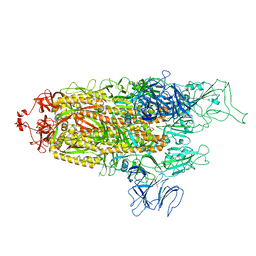 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
6OK8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K127L at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-04-12 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RT1
 
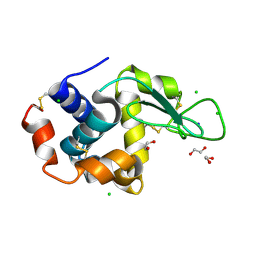 | | Native tetragonal lysozyme - home source data | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Pereira, P.J.B. | | Deposit date: | 2019-05-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | Protein crystals as a key for deciphering macromolecular crowding effects on biological reactions.
Phys Chem Chem Phys, 22, 2020
|
|
8FR6
 
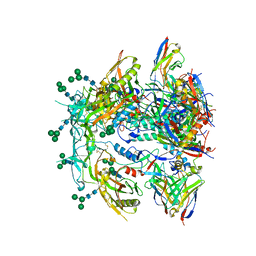 | | Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Wang, S, Kwong, P.D. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8FEZ
 
 | |
6HQD
 
 | | Cytochrome P450-153 from Pseudomonas sp. 19-rlim | | Descriptor: | Cytochrome P450, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-24 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HQW
 
 | |
