1XYM
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | D-glucose, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
8SZB
 
 | |
8PKD
 
 | |
7VAX
 
 | | V1EG of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state1-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAW
 
 | | V1EG domain of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state1-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VB0
 
 | | V1EG domain of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAY
 
 | | V1EG domain of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
3SIO
 
 | | Ac-AChBP ligand binding domain (not including beta 9-10 linker) mutated to human alpha-7 nAChR | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemecz, A, Taylor, P.W. | | Deposit date: | 2011-06-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Creating an alpha-7 nicotinic acetylcholine recognition domain from the
acetylcholine binding protein: crystallographic and ligand selectivity analyses
J.Biol.Chem., 286, 2011
|
|
7UIW
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ0
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIV
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3S2V
 
 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | Descriptor: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
1XYL
 
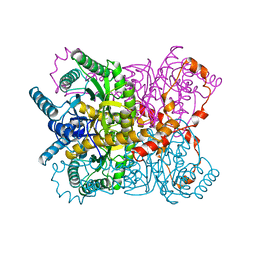 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
3ZFO
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P | | Descriptor: | CHLORIDE ION, HYDROXIDE ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
5W5E
 
 | | Re-refinement of the pyocin tube structure | | Descriptor: | FIIR2 protein | | Authors: | Wang, F, Zheng, W, Taylor, N.M, Guerrero-Ferreira, R.C, Leiman, P.G, Egelman, E.H. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit.
Structure, 25, 2017
|
|
6MPL
 
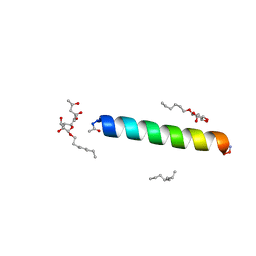 | | Racemic M2-TM I39A crystallized from racemic detergent | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6MPM
 
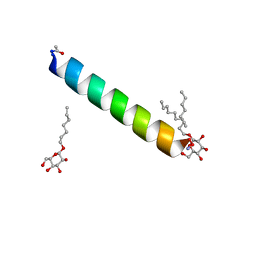 | | Racemic M2-TM I42A crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
4JWY
 
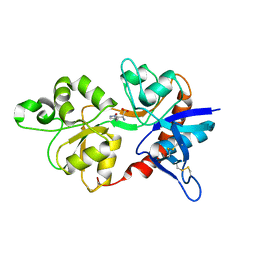 | | GluN2D ligand-binding core in complex with propyl-NHP5G | | Descriptor: | (2R)-amino(1-hydroxy-4-propyl-1H-pyrazol-5-yl)ethanoic acid, Glutamate receptor ionotropic, NMDA 2D | | Authors: | Hansen, K.B, Tajima, N, Risgaard, R, Perszyk, R.E, Jorgensen, L, Vance, K.M, Ogden, K.K, Clausen, R.P, Furukawa, H, Traynelis, S.F. | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-d-aspartate receptors.
Mol.Pharmacol., 84, 2013
|
|
3ORI
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 1) | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Lombana, T.N, Echols, N, Good, M.C, Thomsen, N.D, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3ORO
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 4) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Good, M.C, Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3ORM
 
 | | Mycobacterium tuberculosis PknB kinase domain D76A mutant | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3NHQ
 
 | | The dark Pfr structure of the photosensory core module of P. aeruginosa Bacteriophytochrome | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Yang, X, Ren, Z, Kuk, J, Moffat, K. | | Deposit date: | 2010-06-14 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome.
Nature, 479, 2011
|
|
4G3G
 
 | |
8UB3
 
 | | DpHF7 filament | | Descriptor: | DpHF7 filament | | Authors: | Lynch, E.M, Farrell, D, Shen, H, Kollman, J.M, DiMaio, F, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
