6S6B
 
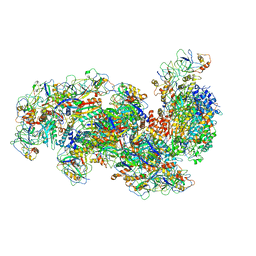 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
7NGQ
 
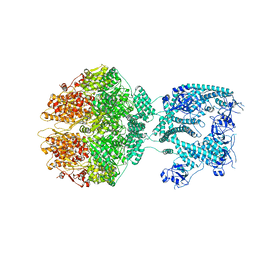 | | Human mitochondrial Lon protease homolog, D2-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Abrahams, J.P, Schmitz, K.A, Maier, T, Schenck, N. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
6FKR
 
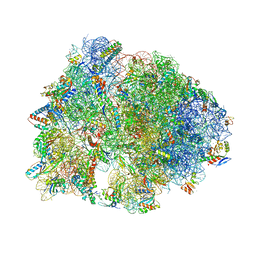 | | Crystal structure of the dolphin proline-rich antimicrobial peptide Tur1A bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16 ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Perebaskine, N, Benincasa, M, Gambato, S, Hofmann, S, Huter, P, Muller, C, Hilpert, K, Innis, C.A, Tossi, A, Wilson, D.N. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-28 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome.
Cell Chem Biol, 25, 2018
|
|
7NGF
 
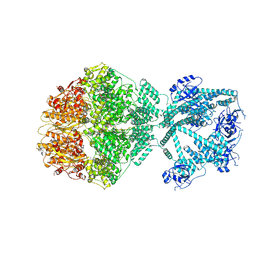 | | P2c-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGP
 
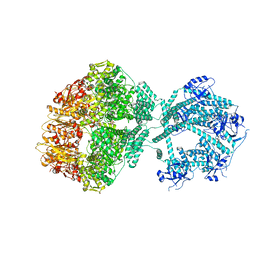 | | D1-state of wild type human mitochondrial LONP1 protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGL
 
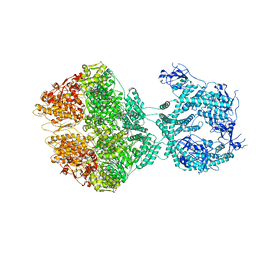 | | R-state of wild type human mitochondrial LONP1 protease bound to endogenous ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7UDA
 
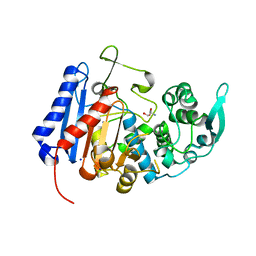 | | Structure of the EstG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase domain-containing protein, SODIUM ION | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2022-03-18 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
7UVQ
 
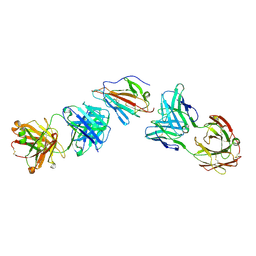 | | Pfs230 domain 1 bound by RUPA-97 and 15C5 Fabs | | Descriptor: | Fab Heavy Chain, Fab Kappa Light Chain, Gametocyte surface protein P230 | | Authors: | Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
7UVI
 
 | | Pfs230 domain 1 bound by RUPA-55 Fab | | Descriptor: | Gametocyte surface protein P230, RUPA-55 Fab heavy chain, RUPA-55 Fab light chain | | Authors: | Ivanochko, D, Newton, J, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
7UVO
 
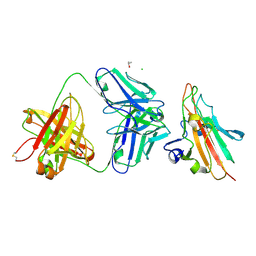 | | Pfs230 domain 1 bound by RUPA-38 Fab | | Descriptor: | CHLORIDE ION, Gametocyte surface protein P230, ISOPROPYL ALCOHOL, ... | | Authors: | Ivanochko, D, Newton, J, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
7UVH
 
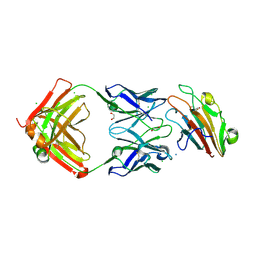 | | Pfs230 domain 1 bound by RUPA-32 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMMONIUM ION, ... | | Authors: | Ivanochko, D, Newton, J, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
7UXL
 
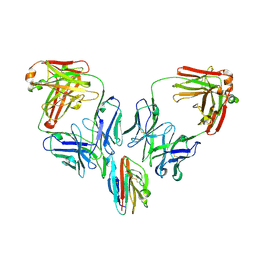 | | Crystal structure of malaria transmission-blocking antigen Pfs48/45-6C variant in complex with human antibodies RUPA-44 and RUPA-29 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gametocyte surface protein P45/48, RUPA-29 Fab Heavy chain, ... | | Authors: | Hailemariam, S, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Highly potent, naturally acquired human monoclonal antibodies against Pfs48/45 block Plasmodium falciparum transmission to mosquitoes.
Immunity, 56, 2023
|
|
7UVS
 
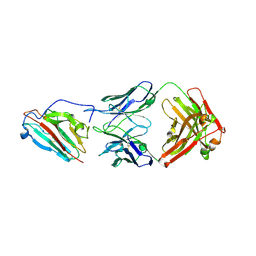 | | Pfs230 domain 1 bound by LMIV230-02 Fab | | Descriptor: | Gametocyte surface protein P230, LMIV230-02 Fab heavy chain, LMIV230-02 Fab light chain, ... | | Authors: | Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
3A5A
 
 | | Crystal structure of a hemoglobin component V from Propsilocerus akamusi (pH5.6 coordinates) | | Descriptor: | Hemoglobin V, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Takagi, T, Shishikura, F. | | Deposit date: | 2009-08-05 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | pH-dependent structural changes in haemoglobin component V from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera)
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7U1B
 
 | | Crystal structure of EstG in complex with tantalum cluster | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, Beta-lactamase domain-containing protein, ... | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2022-02-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
7U1C
 
 | | Structure of EstG crystalized with SO4 and Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase domain-containing protein, SODIUM ION, ... | | Authors: | Gabelli, S.B, Chen, Z. | | Deposit date: | 2022-02-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
3A5B
 
 | | Crystal structure of a hemoglobin component V from Propsilocerus akamusi (pH6.5 coordinates) | | Descriptor: | Hemoglobin V, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Takagi, T, Shishikura, F. | | Deposit date: | 2009-08-05 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | pH-dependent structural changes in haemoglobin component V from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera)
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A5G
 
 | | Crystal structure of a hemoglobin component V from Propsilocerus akamusi (pH7.0 coordinates) | | Descriptor: | CARBON MONOXIDE, Hemoglobin V, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Takagi, T, Shishikura, F. | | Deposit date: | 2009-08-06 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | pH-dependent structural changes in haemoglobin component V from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera)
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A9M
 
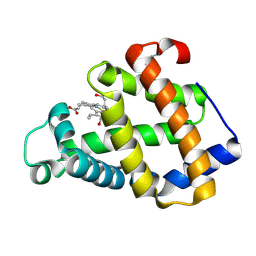 | | Crystal structure of a hemoglobin component V from Propsilocerus akamusi (pH9.0 coordinates) | | Descriptor: | CARBON MONOXIDE, Hemoglobin V, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Takagi, T, Shishikura, F. | | Deposit date: | 2009-10-30 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH-dependent structural changes in haemoglobin component V from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera)
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XYK
 
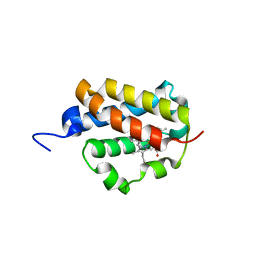 | | Group II 2-on-2 Hemoglobin from the Plant Pathogen Agrobacterium tumefaciens | | Descriptor: | 2-ON-2 HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Nardini, M, LaBarre, M, Richard, C, Wittenberg, J.B, Wittenberg, B.A, Guertin, M, Bolognesi, M. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of a Group II 2/2 Hemoglobin from the Plant Pathogen Agrobacterium Tumefaciens.
Biochim.Biophys.Acta, 1814, 2011
|
|
2ZWJ
 
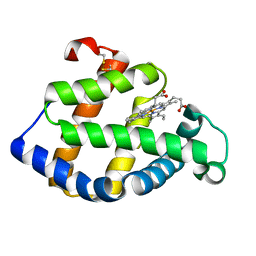 | | Crystal structure of a hemoglobin component V from Propsilocerus akamusi (pH4.6 coordinates) | | Descriptor: | Hemoglobin V, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Takagi, T, Shishikura, F. | | Deposit date: | 2008-12-13 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | pH-dependent structural changes in haemoglobin component V from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera)
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7U8E
 
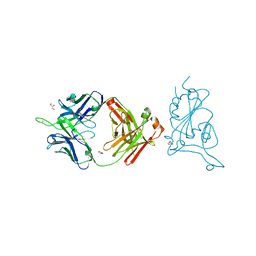 | |
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
