2ML9
 
 | |
2MLA
 
 | |
2MLB
 
 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | Descriptor: | redesigned ubiquitin | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MLD
 
 | |
2MLE
 
 | | NMR structure of the C-domain of troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Baryshnikova, O.K, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
2MLF
 
 | | NMR structure of the dilated cardiomyopathy mutation G159D in troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Baryshnikova, O.K, Robertson, I.M, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
2MLG
 
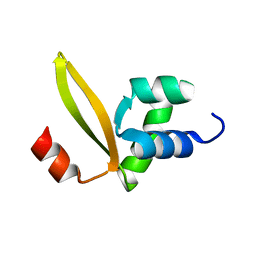 | | Stf76 from the Sulfolobus islandicus plasmid-virus pSSVx | | Descriptor: | Sulfolobus transcription factor 76 aminoacid protein, Stf76 | | Authors: | Farina, B, Russo, L, Fattorusso, R. | | Deposit date: | 2014-02-27 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of Stf76 from the Sulfolobus islandicus plasmid-virus pSSVx: a novel peculiar member of the winged helix-turn-helix transcription factor family.
Nucleic Acids Res., 42, 2014
|
|
2MLH
 
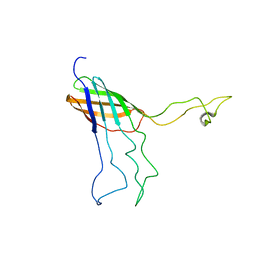 | | NMR Solution Structure of Opa60 from N. Gonorrhoeae in FC-12 Micelles | | Descriptor: | Opacity protein opA60 | | Authors: | Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M, Columbus, L. | | Deposit date: | 2014-02-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the neisserial outer membrane protein opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
2MLI
 
 | |
2MLJ
 
 | |
2MLK
 
 | | Three-dimensional structure of the C-terminal DNA-binding domain of RstA protein from Klebsiella pneumoniae | | Descriptor: | RstA | | Authors: | Fang, P, Chen, S, Cheng, Y, Chang, C, Yu, T, Huang, T. | | Deposit date: | 2014-03-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
2MLL
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
2MLM
 
 | | Solution structure of sortase A from S. aureus in complex with benzo[d]isothiazol-3-one based inhibitor | | Descriptor: | N-{2-oxo-2-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylamino]ethyl}-2-sulfanylbenzamide, Sortase family protein | | Authors: | Jaudzems, K, Zhulenkovs, D, Leonchiks, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-11-19 | | Last modified: | 2018-06-27 | | Method: | SOLUTION NMR | | Cite: | Discovery and structure-activity relationship studies of irreversible benzisothiazolinone-based inhibitors against Staphylococcus aureus sortase A transpeptidase.
Bioorg.Med.Chem., 22, 2014
|
|
2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2MLP
 
 | | MICROCIN LEADER PEPTIDE FROM E. COLI, NMR, 25 STRUCTURES | | Descriptor: | MCBA PROPEPTIDE | | Authors: | Kim, S, Sinha Roy, R, Walsh, C.T, Baleja, J.D. | | Deposit date: | 1998-01-21 | | Release date: | 1998-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26.
Chem.Biol., 5, 1998
|
|
2MLQ
 
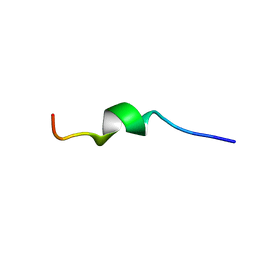 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2MLR
 
 | | Membrane Bilayer complex with Matrix Metalloproteinase-12 at its Alpha-face | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Koppisetti, R.K, Fulcher, Y.G, Prior, S.H, Lenoir, M, Overduin, M, Van Doren, S.R. | | Deposit date: | 2014-03-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12.
Nat Commun, 5, 2014
|
|
2MLS
 
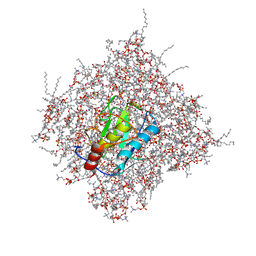 | | Membrane Bilayer complex with Matrix Metalloproteinase-12 at its Beta-face | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Koppisetti, R.K, Fulcher, Y.G, Prior, S.H, Lenoir, M, Overduin, M, Van Doren, S.R. | | Deposit date: | 2014-03-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12.
Nat Commun, 5, 2014
|
|
2MLT
 
 | |
2MLU
 
 | |
2MLV
 
 | |
2MLW
 
 | | New Cyt-like delta-endotoxins from Dickeya dadantii - CytC protein | | Descriptor: | Type-1Ba cytolytic delta-endotoxin | | Authors: | Loth, K, Costechareyre, D, Effantin, G, Rahbe, Y, Condemine, G, Landon, C, Da Silva, P. | | Deposit date: | 2014-03-05 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | New Cyt-like delta-endotoxins from Dickeya dadantii: structure and aphicidal activity.
Sci Rep, 5, 2015
|
|
2MLX
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA220-310 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2MLY
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA1-150 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2MLZ
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA365-471 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
