2MHG
 
 | |
2MHH
 
 | | Solution structure of a EF-hand domain from sea urchin polycystin-2 | | Descriptor: | Polycystic kidney disease protein 2 | | Authors: | Kuo, I.Y, Keeler, C, Corbin, R, Celic, A, Petri, E.T, Hodsdon, M.E, Ehrlich, B.E. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The number and location of EF hand motifs dictates the calcium dependence of polycystin-2 function.
Faseb J., 28, 2014
|
|
2MHI
 
 | |
2MHJ
 
 | |
2MHK
 
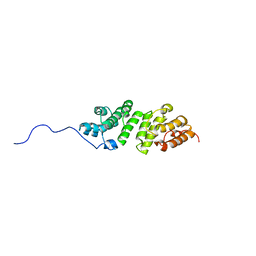 | | E. coli LpoA N-terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Jean, N.L, Bougault, C, Lodge, A, Derouaux, A, Callens, G, Egan, A, Lewis, R.J, Vollmer, W, Simorre, J. | | Deposit date: | 2013-11-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Elongated Structure of the Outer-Membrane Activator of Peptidoglycan Synthesis LpoA: Implications for PBP1A Stimulation.
Structure, 22, 2014
|
|
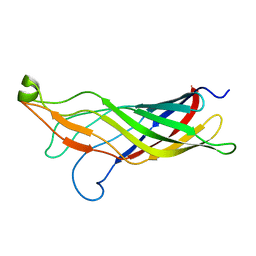
2MHL
 
 | |
2MHM
 
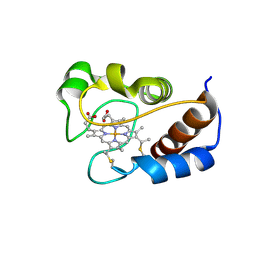 | | Solution structure of cytochrome c Y67H | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W.X, Wang, Z.H, Yang, Z.Z, Ying, T.L, Wu, H.M, Tan, X.S, Cao, C.Y, Huang, Z.X. | | Deposit date: | 2013-11-26 | | Release date: | 2014-10-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cytochrome c Y67H Mutant to Function as a Peroxidase
Plos One, 9, 2014
|
|
2MHN
 
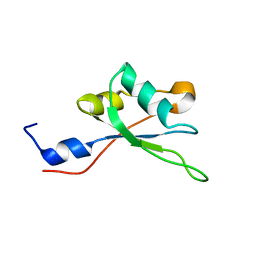 | |
2MHO
 
 | |
2MHP
 
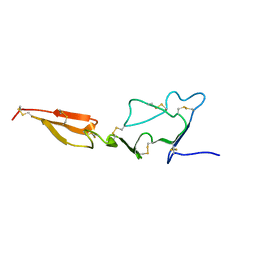 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
2MHQ
 
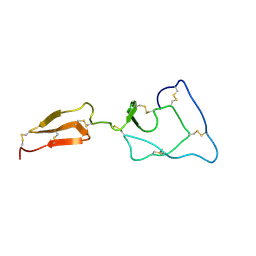 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
2MHR
 
 | |
2MHS
 
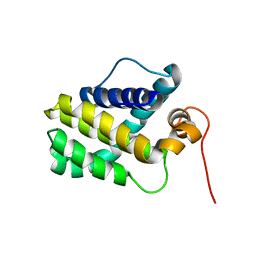 | | NMR Structure of human Mcl-1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, G, Poppe, L, Aoki, K, Yamane, H, Lewis, J, Szyperski, T. | | Deposit date: | 2013-12-04 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Quality NMR Structure of Human Anti-Apoptotic Protein Domain Mcl-1(171-327) for Cancer Drug Design.
Plos One, 9, 2014
|
|
2MHU
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
2MHV
 
 | | Solution Structure of Penicillium Antifungal Protein PAF | | Descriptor: | Antifungal protein | | Authors: | Fizil, A, Batta, G. | | Deposit date: | 2013-12-05 | | Release date: | 2014-12-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | "Invisible" Conformers of an Antifungal Disulfide Protein Revealed by Constrained Cold and Heat Unfolding, CEST-NMR Experiments, and Molecular Dynamics Calculations.
Chemistry, 21, 2015
|
|
2MHW
 
 | | The solution NMR structure of maximin-4 in SDS micelles | | Descriptor: | Antimicrobial peptide | | Authors: | Toke, O, Banoczi, Z, Kiraly, P, Heinzmann, R, Burck, J, Ulrich, A.S, Hudecz, F. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A kinked antimicrobial peptide from Bombina maxima. I. Three-dimensional structure determined by NMR in membrane-mimicking environments.
Eur.Biophys.J., 40, 2011
|
|
2MHX
 
 | | Structure of Exocyclic R,R N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(RBD)P*AP*GP*AP*AP*G)-3'), 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2MHY
 
 | | Structure determination of the salamander courtship pheromone Plethodontid Modulating Factor | | Descriptor: | Plethodontid modulating factor | | Authors: | Wilburn, D.B, Bowen, K.E, Doty, K.A, Arumugam, S, Lane, A.N, Feldhoff, P.W, Feldhoff, R.C. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the evolution of a sexy protein: novel topology and restricted backbone flexibility in a hypervariable pheromone from the red-legged salamander, Plethodon shermani.
Plos One, 9, 2014
|
|
2MHZ
 
 | | Structure of Exocyclic S,S N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(SDE)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2MI0
 
 | | NMR structure of the I-V kissing-loop interaction of the Neurospora VS ribozyme | | Descriptor: | 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3' | | Authors: | Bouchard, P, Legault, P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate recognition by the neurospora varkud satellite ribozyme: importance of u-turns at the kissing-loop junction.
Biochemistry, 53, 2014
|
|
2MI1
 
 | |
2MI2
 
 | |
2MI5
 
 | |
2MI6
 
 | |
2MI7
 
 | | Solution NMR structure of alpha3Y | | Descriptor: | de novo protein a3Y | | Authors: | Glover, S.D, Jorge, C, Liang, L, Valentine, K.G, Hammarstrom, L, Tommos, C. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Photochemical tyrosine oxidation in the structurally well-defined alpha 3Y protein: proton-coupled electron transfer and a long-lived tyrosine radical.
J.Am.Chem.Soc., 136, 2014
|
|
