7ZRT
 
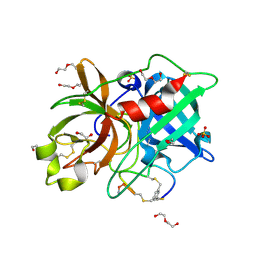 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970
To Be Published
|
|
8YSW
 
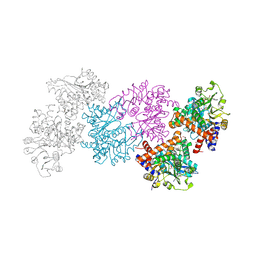 | | phosphinothricin dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Glutamate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Xue, Y.P, Cheng, F, Zhou, S.P, Xu, J.M, Jin, L.Q, Ma, C.J, Zheng, Y.G. | | Deposit date: | 2024-03-23 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of phosphinothricin dehydrogenase with NAD at 2.6 Angstorms resolution
To Be Published
|
|
5JNW
 
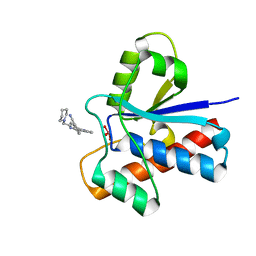 | | Crystal structure of bovine low molecular weight protein tyrosine phosphatase (LMPTP) mutant (W49Y N50E) complexed with vanadate and uncompetitive inhibitor | | Descriptor: | 2-(4-{[3-(piperidin-1-yl)propyl]amino}quinolin-2-yl)benzonitrile, Low molecular weight phosphotyrosine protein phosphatase, VANADATE ION | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
6GUH
 
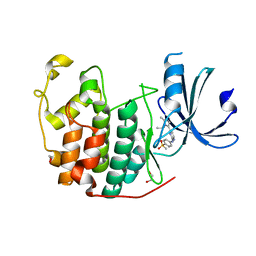 | | CDK2 in complex with AZD5438 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
7OPG
 
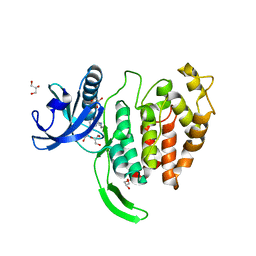 | | Crystal structure of CLK1 in complex with compound 2 (CC513) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[2-(propylamino)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]phenol, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Routier, S, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of CLK1 in complex with compound 2 (CC513)
To Be Published
|
|
8ZD8
 
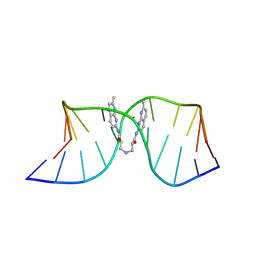 | | NMR structure of the (CGG-dsDNA:ND=) 1:1 complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3'), ~{N}-(7-methyl-1,8-naphthyridin-2-yl)-3-[[3-[(7-methyl-1,8-naphthyridin-2-yl)amino]-3-oxidanylidene-propyl]amino]propanamide | | Authors: | Sakurabayashi, S, Furuita, K, Yamada, T, Nomura, M, Nakatani, K, Kojima, C. | | Deposit date: | 2024-05-01 | | Release date: | 2025-04-30 | | Last modified: | 2025-11-12 | | Method: | SOLUTION NMR | | Cite: | NMR-Based Rational Drug Design of G:G Mismatch DNA Binding Ligand Trapping Transient Complex via Disruption of a Key Allosteric Interaction.
J.Am.Chem.Soc., 147, 2025
|
|
1F8E
 
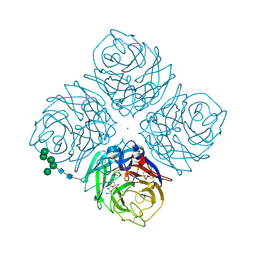 | | Native Influenza Neuraminidase in Complex with 4,9-diamino-2-deoxy-2,3-dehydro-N-acetyl-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,9-AMINO-2,4-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
9KKP
 
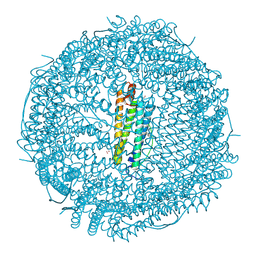 | | Crystal structure of Horse spleen L-ferritin mutant (E53F/E56F/E57F/R59F/E60F/E63F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
7TWE
 
 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DUF1479 domain-containing protein, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-07 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid
To Be Published
|
|
7ONB
 
 | | Structure of the U2 5' module of the A3'-SSA complex | | Descriptor: | MINX, PHD finger-like domain-containing protein 5A, RNU2, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
6YI4
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Zak, K.M, Zhou, R.X, Softley, C.A, Bostock, M.J, Sattler, M, Popowicz, G.M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion
Not published
|
|
5IUI
 
 | | Crystal Structure of Anaplastic Lyphoma Kinase (ALK) in complex with 4 | | Descriptor: | ALK tyrosine kinase receptor, N-[3-(4-amino-3-methylphenyl)-1H-pyrazol-5-yl]-4-[(4-methylpiperazin-1-yl)methyl]benzamide | | Authors: | Tu, C.H, Wu, S.Y. | | Deposit date: | 2016-03-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Pyrazolylamine Derivatives Reveal the Conformational Switching between Type I and Type II Binding Modes of Anaplastic Lymphoma Kinase (ALK).
J.Med.Chem., 59, 2016
|
|
6W6D
 
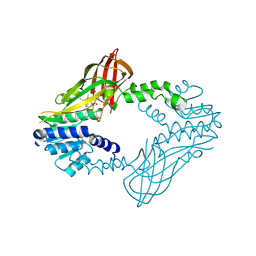 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
7QRX
 
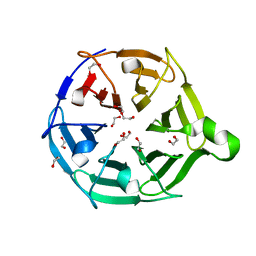 | |
7QT6
 
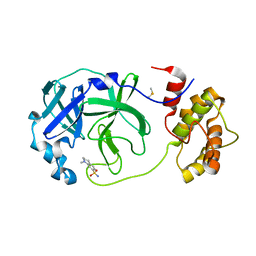 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z1367324110 | | Descriptor: | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8AHE
 
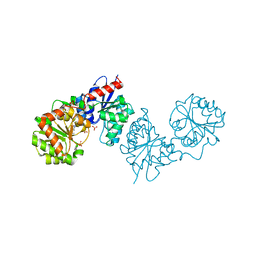 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8FTJ
 
 | | Crystal structure of human NEIL1 (P2G (242K) C(delta)100) glycosylase bound to DNA duplex containing urea | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*TP*CP*CP*AP*UDV*GP*TP*CP*TP*AP*CP)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*GP*G)-3'), ... | | Authors: | Tomar, R, Sharma, P, Harp, J.M, Egli, M, Stone, M.P. | | Deposit date: | 2023-01-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.
Nucleic Acids Res., 51, 2023
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
7QS1
 
 | |
8FLT
 
 | | Human PTH1R in complex with M-PTH(1-14) and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
5IXV
 
 | |
6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
8U06
 
 | | Imine reductase RedE bound with NADP+ and arcyriaflavin A (primary site) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arcyriaflavin A, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-08-28 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An imine reductase that captures reactive intermediates in the biosynthesis of the indolocarbazole reductasporine.
J.Biol.Chem., 300, 2024
|
|
7OXZ
 
 | | VDR complex with a side-chain hydroxylated derivative of lithocholic acid | | Descriptor: | (3R,6R)-6-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]heptane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-06-23 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis and evaluation of side-chain hydroxylated derivatives of lithocholic acid as potent agonists of the vitamin D receptor (VDR).
Bioorg.Chem., 115, 2021
|
|
7OY4
 
 | | VDR complex of a side-chain hydroxylated derivatives of lithocholic acid | | Descriptor: | (3S,6R)-6-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]heptane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-06-23 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and evaluation of side-chain hydroxylated derivatives of lithocholic acid as potent agonists of the vitamin D receptor (VDR).
Bioorg.Chem., 115, 2021
|
|
