7SFE
 
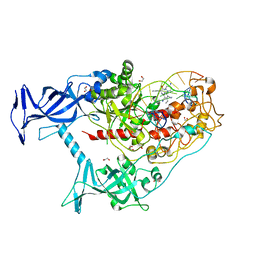 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830334A | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]amino}-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
4US6
 
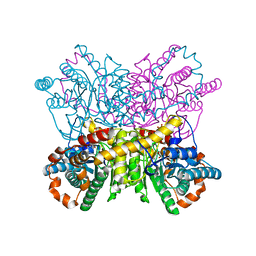 | | New Crystal Form of Glucose Isomerase Grown in Short Peptide Supramolecular Hydrogels | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Diaz-Mochon, J.J, Alvarez de Cienfuegos, L. | | Deposit date: | 2014-07-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Influence of the Chirality of Short Peptide Supramolecular Hydrogels in Protein Crystallogenesis.
Chem.Commun.(Camb.), 51, 2015
|
|
9I0U
 
 | | Structure of human PD-L1 in complex with clinically evaluated inhibitor | | Descriptor: | (5~{S})-5-[[[5-[2-chloranyl-3-[2-chloranyl-3-[6-methoxy-5-[[[(2~{S})-5-oxidanylidenepyrrolidin-2-yl]methylamino]methyl]pyrazin-2-yl]phenyl]phenyl]-3-methoxy-pyrazin-2-yl]methylamino]methyl]pyrrolidin-2-one, GLYCEROL, Programmed cell death 1 ligand 1 | | Authors: | Golebiowska-Mendroch, K, Slota, A, Plewka, J, Magiera-Mularz, K. | | Deposit date: | 2025-01-15 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Characterization of Clinically Evaluated Small-Molecule Inhibitors of PD-L1 for Immunotherapy.
Acs Med.Chem.Lett., 16, 2025
|
|
9HRT
 
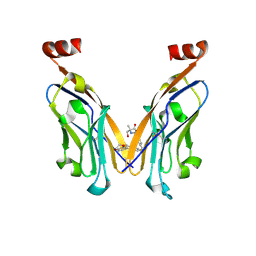 | | Structure of human PD-L1 in complex with clinically evaluated inhibitor | | Descriptor: | (2~{R})-2-[[3-[(~{E})-2-[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]ethenyl]-4-(trifluoromethyl)phenyl]methylamino]-2-methyl-3-oxidanyl-propanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Slota, A, Golebiowska-Mendroch, K, Plewka, J, Magiera-Mularz, K. | | Deposit date: | 2024-12-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Clinically Evaluated Small-Molecule Inhibitors of PD-L1 for Immunotherapy.
Acs Med.Chem.Lett., 16, 2025
|
|
7RYN
 
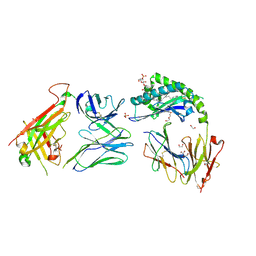 | | CD1a-sulfatide-gdTCR complex | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
4WD6
 
 | |
7S6H
 
 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
9HZ5
 
 | | Pre-clinical characterization of novel multi-client inhibitors of Sec61 with broad anti-tumor activity | | Descriptor: | 4-{(3S)-9-(cyclohexylmethyl)-5-[(3R,5R)-4-(3-fluoro-5-methoxyphenyl)-3,5-dimethylpiperazine-1-sulfonyl]-3-methyl-1,5,9-triazacyclododecane-1-sulfonyl}-N,N-dimethylaniline, Protein transport protein Sec61 subunit alpha, Protein transport protein Sec61 subunit beta, ... | | Authors: | Shahid, R, Paavilainen, V.O. | | Deposit date: | 2025-01-13 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Preclinical characterization of novel multi-client inhibitors of Sec61 with broad antitumor activity.
J.Pharmacol.Exp.Ther., 392, 2025
|
|
7Z5J
 
 | | The molybdenum storage protein loaded with tungstate | | Descriptor: | 1,1,3,3,5,7,7,9,11,15,15-undecakis($l^{1}-oxidanyl)-2$l^{4},4$l^{3},6$l^{5},8,10,12,14,16,17,18,19$l^{3},20,21,22,23-pentadecaoxa-1$l^{6},3$l^{6},5$l^{6},7$l^{6},9$l^{6},11$l^{6},13$l^{6},15$l^{6}-octatungstapentadecacyclo[7.7.1.1^{1,13}.1^{3,5}.1^{3,15}.1^{5,7}.1^{5,11}.1^{7,11}.0^{2,13}.0^{2,15}.0^{4,13}.0^{6,9}.0^{6,11}.0^{6,13}.0^{9,19}]tricosane, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ermler, U, Aziz, I, Kaltwasser, S, Kayastha, K, Vonck, J. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The molybdenum storage protein forms and deposits distinct polynuclear tungsten oxygen aggregates.
J.Inorg.Biochem., 234, 2022
|
|
9I6Y
 
 | | 14-3-3sigma binding to the ERa peptide and compound 1 | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[4-[3-[(phenylmethyl)amino]imidazo[1,2-a]pyridin-2-yl]phenyl]ethanamide, CALCIUM ION, ... | | Authors: | Pennings, M.A.M, Arkin, M.R. | | Deposit date: | 2025-01-31 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Scaffold-hopping for molecular glues targeting the 14-3-3/ER alpha complex.
Nat Commun, 16, 2025
|
|
8JTN
 
 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 2-propyl-2-azoniatricyclo[7.3.0.0^{3,7}]dodeca-1(9),2,7-trien-8-amine, Tudor domain-containing protein 3 | | Authors: | Chen, M, Wang, Z, Li, W, Shang, X, Liu, Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Tudor domain of TDRD3 in complex with a small molecule antagonist.
Biochim Biophys Acta Gene Regul Mech, 1866, 2023
|
|
8JPF
 
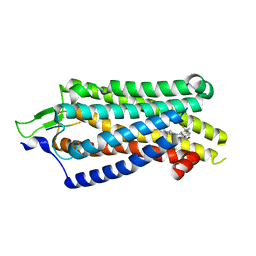 | | Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, NTS, Neurotensin receptor type 1 | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
4W9U
 
 | |
8JWZ
 
 | | Crystal structure of A2AR-T4L in complex with AB928 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
9HT2
 
 | | A novel bottom-up approach to find lead-compounds in billion-sized libraries | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Bromodomain-containing protein 4, methyl 2,2,6,6-tetramethyl-4-[[2-[(5-methyl-1,3,4-thiadiazol-2-yl)amino]-2-oxidanylidene-ethyl]amino]oxane-4-carboxylate | | Authors: | Ruiz Carrillo, D, Garcia Alai, M, Barril, X, Serrano-Morras, A, Bertran-Mostazo, A, Juarez-Jimenez, J, Defelipe, L. | | Deposit date: | 2024-12-19 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A bottom-up approach to find lead compounds in expansive chemical spaces.
Commun Chem, 8, 2025
|
|
7SAB
 
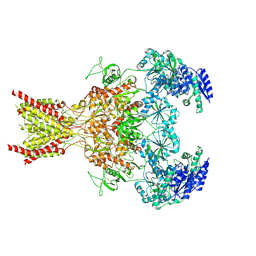 | | Phencyclidine-bound GluN1a-GluN2B NMDA receptors | | Descriptor: | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
9HWS
 
 | | Crystal structure of the Keap1 Kelch domain in complex with the small molecule UCAB#909 at 1.61 Angstrom resolution | | Descriptor: | 3-[7-[2-(cycloheptylamino)-2-oxidanylidene-ethoxy]naphthalen-2-yl]propanoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2025-01-06 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of Novel High-affinity, Selective, and Anti-inflammatory Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9IC2
 
 | | Structure of AMPK-alpha2-kinase domain bound to BAY-3827 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ahangar, M.S, Zeqiraj, E, Fraguas Bringas, C, Sakamoto, K. | | Deposit date: | 2025-02-14 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of AMPK-alpha2-kinase domain bound to BAY-3827
To Be Published
|
|
9HWQ
 
 | | Crystal structure of the Keap1 Kelch domain in complex with the XChem fragment Z19735904 at 1.14 Angstrom resolution. | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2025-01-06 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Fragment-Based Drug Discovery of Novel High-affinity, Selective, and Anti-inflammatory Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9HEQ
 
 | |
9HEO
 
 | |
4V33
 
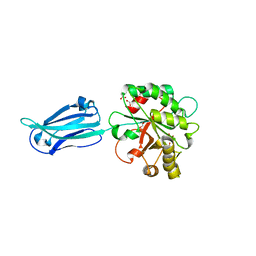 | | Crystal structure of the putative polysaccharide deacetylase BA0330 from bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, POLYSACCHARIDE DEACETYLASE-LIKE PROTEIN, ... | | Authors: | Giastas, P, Andreou, A, Bouriotis, V, Eliopoulos, E. | | Deposit date: | 2014-10-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Two Putative Polysaccharide Deacetylases are Required for Osmotic Stability and Cell Shape Maintenance in Bacillus Anthracis.
J.Biol.Chem., 290, 2015
|
|
7SHV
 
 | | Crystal structure of BRAF kinase domain bound to GDC0879 | | Descriptor: | 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Kung, J.E, Sudhamsu, J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Clearing the Path to Rapid High-Quality Protein Purification
To Be Published
|
|
9I97
 
 | | Cryo-EM structure of Shigella flexneri LptDE in complex with a Bicyclic Peptide binder (Compound 12) | | Descriptor: | 1-[4,7-bis(2-chloranylethanoyl)-1,4,7-triazonan-1-yl]-2-chloranyl-ethanone, Compound 12 - Bicyclic Peptide Binder, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Allyjaun, S, Dunbar, E, Hardwick, S.W, Chirgadze, D.Y, Hubbard, J, van den Berg, B, Newman, H. | | Deposit date: | 2025-02-06 | | Release date: | 2025-10-15 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | High-Throughput Identification and Characterization of LptDE-Binding Bicycle Peptides Using Phage Display and Cryo-EM.
J.Med.Chem., 68, 2025
|
|
9I96
 
 | | Cryo-EM structure of Shigella flexneri LptDE bound by a Bicyclic peptide molecule (Compound 5) | | Descriptor: | 1-[3,5-bis(2-chloranylethanoyl)-1,3,5-triazinan-1-yl]-2-chloranyl-ethanone, Compound 5 - Bicyclic Peptide binder, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Allyjaun, S, Newman, H, Dunbar, E, Hardwick, S.W, Chirgadze, D.Y, van den Berg, B, Hubbard, J. | | Deposit date: | 2025-02-06 | | Release date: | 2025-10-15 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | High-Throughput Identification and Characterization of LptDE-Binding Bicycle Peptides Using Phage Display and Cryo-EM.
J.Med.Chem., 68, 2025
|
|
