7L89
 
 | | BG505 SOSIP MD39 in complex with the polyclonal Fab pAbC-4 from animal Rh.32034 (Wk26 time point) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP MD39 - gp120, ... | | Authors: | Antanasijevic, A, Sewall, L.M, Rantalainen, K, Perrett, H.R, Ward, A.B. | | Deposit date: | 2020-12-31 | | Release date: | 2021-08-04 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM.
Nat Commun, 12, 2021
|
|
5UEK
 
 | |
5USL
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv304 | | Descriptor: | Fab vv304 Heavy chain, Fab vv304 Light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
5USH
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv66 | | Descriptor: | Fab vv66 heavy chain, Fab vv66 light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
6SRV
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6CR1
 
 | | adalimumab EFab | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heavy chain of adalimumab EFab (VH-IgE CH2), Light chain of adalimumab EFab (VL-IgE CH2), ... | | Authors: | Arndt, J.W. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | EFab domain substitution as a solution to the light-chain pairing problem of bispecific antibodies.
MAbs, 10, 2018
|
|
8GV2
 
 | | Crystal structure of anti-FX IgG fab without FAST-Ig mutations | | Descriptor: | 1,2-ETHANEDIOL, Anti-factor X IgG fab heavy chain, Anti-factor X IgG fab light chain | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
8GV1
 
 | | Crystal structure of anti-FX IgG fab with FAST-Ig mutations | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Anti-factor X IgG fab heavy chain, ... | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.186 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
8GUZ
 
 | | Crystal structure of anti-FIXa IgG fab with FAST-Ig mutations | | Descriptor: | 1,2-ETHANEDIOL, Anti-factor IXa IgG fab heavy chain, Anti-factor IXa IgG fab light chain | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
8GV0
 
 | | Crystal structure of anti-FIXa IgG fab without FAST-Ig mutations | | Descriptor: | Anti-factor IXa IgG fab heavy chain, Anti-factor IXa IgG fab light chain | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
6TUL
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021177 | | Descriptor: | D-MALATE, Fab C0021177 heavy chain (IgG1), Fab C0021177 light chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6U50
 
 | | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) | | Authors: | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | Deposit date: | 2019-08-26 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
6U53
 
 | |
7RAH
 
 | | Adenylate cyclase toxin RTX domain fragment bound to M1H5 Fab and M2B10 Fab | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA,Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, M1H5 Fab Heavy Chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2021-07-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for antibody binding to adenylate cyclase toxin reveals RTX linkers as neutralization-sensitive epitopes.
Plos Pathog., 17, 2021
|
|
6UYN
 
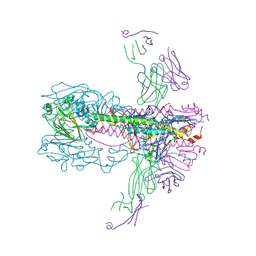 | |
6O1D
 
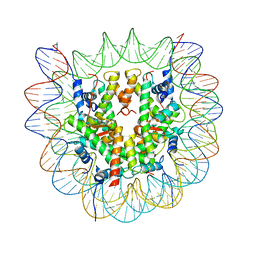 | |
7JWQ
 
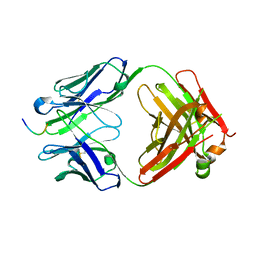 | |
7JWP
 
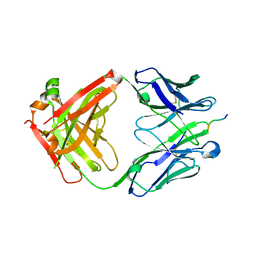 | | Fab CJ11 in complex IL-18 peptide liberated by Caspase cleavage | | Descriptor: | Fab CJ11 Heavy chain, Fab CJ11 Light chain, IL-18 peptide | | Authors: | Payandeh, J, Ho, H. | | Deposit date: | 2020-08-26 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of a caspase cleavage motif antibody reveals insights into noncanonical inflammasome function.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5Z3I
 
 | |
7K93
 
 | |
6VGR
 
 | |
5Z37
 
 | |
7WOB
 
 | | SARS-CoV-2 Spike in complex with IgG 553-60 (2-up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOG
 
 | | SARS-CoV-2 Omicron S monomer complexed with 553-49 | | Descriptor: | 553-49 VH, 553-49 VL, Spike protein S1 | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
5Z3J
 
 | |
