6HHC
 
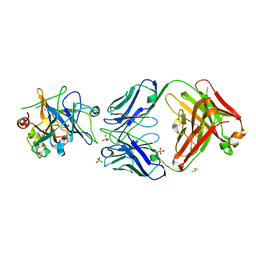 | | Allosteric Inhibition as a new mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated form of Coagulation Factor XI | | Descriptor: | Coagulation factor XI, DIMETHYL SULFOXIDE, FXIA ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Schaefer, M, Buchmueller, A, Dittmer, F, Strassburger, J, Wilmen, A. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Inhibition as a New Mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated Form of Coagulation Factor XI.
J.Mol.Biol., 431, 2019
|
|
3DUY
 
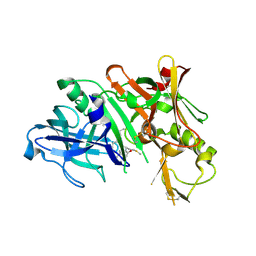 | | Crystal structure of human beta-secretase in complex with NVP-AFJ144 | | Descriptor: | (2R,4S,5S)-N-butyl-4-hydroxy-2,7-dimethyl-5-{[N-(4-methylpentanoyl)-L-methionyl]amino}octanamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2008-07-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based design and synthesis of macrocyclic peptidomimetic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6HMQ
 
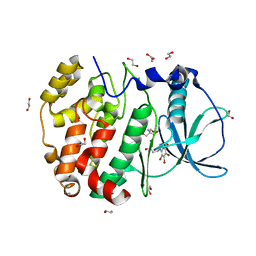 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BENZOTRIAZOLE-TYPE INHIBITOR MB002 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
3DVS
 
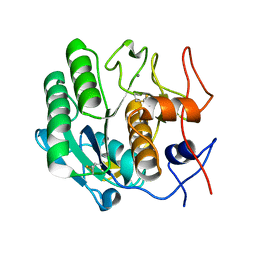 | |
5E7W
 
 | | X-ray Structure of Human Recombinant 2Zn insulin at 0.92 Angstrom | | Descriptor: | ACETATE ION, Insulin, N-PROPANOL, ... | | Authors: | Lisgarten, D.R, Naylor, C.E, Palmer, R.A, Lobley, C.M.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.9519 Å) | | Cite: | Ultra-high resolution X-ray structures of two forms of human recombinant insulin at 100 K.
Chem Cent J, 11, 2017
|
|
6SCV
 
 | | Endothiapepsin in complex with ligand 69 | | Descriptor: | Endothiapepsin, GLYCEROL, [(~{R})-cyclohexyl-[1-(2-phenylethyl)-1,2,3,4-tetrazol-5-yl]methyl]diazane | | Authors: | Magari, F, Heine, A, Klebe, G. | | Deposit date: | 2019-07-25 | | Release date: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Endothiapepsin in complex with ligand 69
To Be Published
|
|
5E85
 
 | | isolated SBD of BiP | | Descriptor: | 78 kDa glucose-regulated protein, SULFATE ION | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
3DJQ
 
 | | Bovine Seminal Ribonuclease- Uridine 5' diphosphate complex | | Descriptor: | Seminal ribonuclease, URIDINE-5'-DIPHOSPHATE | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
6HOP
 
 | |
3DJV
 
 | | Bovine Seminal Ribonuclease- cytidine 3' phosphate complex | | Descriptor: | CYTIDINE-3'-MONOPHOSPHATE, Seminal ribonuclease | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
5E8W
 
 | |
5E95
 
 | | Crystal Structure of Mb(NS1)/H-Ras Complex | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eguchi, R.R, Sha, F, Gupta, A, Koide, A, Koide, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Inhibition of RAS function through targeting an allosteric regulatory site.
Nat. Chem. Biol., 13, 2017
|
|
6THA
 
 | | Crystal structure of human sugar transporter GLUT1 (SLC2A1) in the inward conformation | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Solute carrier family 2, ... | | Authors: | Pedersen, B.P, Paulsen, P.A, Custodio, T.F. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of GLUT1 to GLUT3 reveal transport regulation mechanism in sugar porter family.
Life Sci Alliance, 4, 2021
|
|
6HQD
 
 | | Cytochrome P450-153 from Pseudomonas sp. 19-rlim | | Descriptor: | Cytochrome P450, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-24 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
5EAC
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor R-tebuconazole | | Descriptor: | (3~{R})-1-(4-chlorophenyl)-4,4-dimethyl-3-(1,2,4-triazol-1-ylmethyl)pentan-3-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tyndall, J.D.A, Sabherwal, M, Sagatova, A.A, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
6HQN
 
 | | Crystal structure of GcoA F169L bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQW
 
 | |
5E4G
 
 | | Crystal structure of human growth differentiation factor 11 (GDF-11) | | Descriptor: | Growth/differentiation factor 11, TETRAETHYLENE GLYCOL | | Authors: | Padyana, A.K, Vaidialingam, B, Hayes, D.B, Gupta, P, Franti, M, Farrow, N.A. | | Deposit date: | 2015-10-06 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human GDF11.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6HMB
 
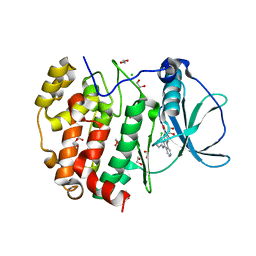 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 Gene product) IN COMPLEX WITH the inhibitor CX-4945 (Silmitasertib) | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
5E55
 
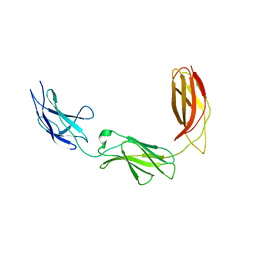 | |
3DND
 
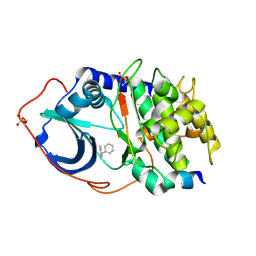 | | cAMP-dependent protein kinase PKA catalytic subunit with PKI-5-24 | | Descriptor: | 5-benzyl-1,3-thiazol-2-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2008-07-02 | | Release date: | 2009-06-23 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallography-independent determination of ligand binding modes
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
6HOU
 
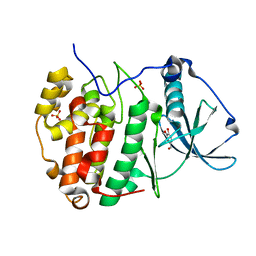 | | Human protein kinase CK2 alpha in complex with vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Battistutta, R, Lolli, G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and cellular mechanism of protein kinase CK2 inhibition by deceptive curcumin.
Febs J., 287, 2020
|
|
6T25
 
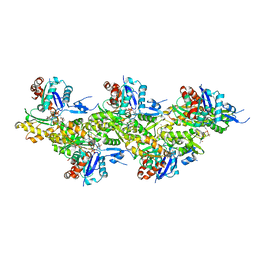 | | Cryo-EM structure of phalloidin-Alexa Flour-546-stabilized F-actin (copolymerized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Merino, F, Raunser, S. | | Deposit date: | 2019-10-07 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Structure, 28, 2020
|
|
3DEP
 
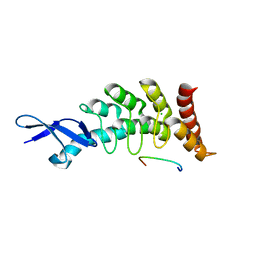 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
5DNF
 
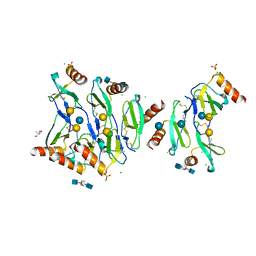 | | Crystal structure of CC chemokine 5 (CCL5) oligomer in complex with heparin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, C-C motif chemokine 5, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
