2CCK
 
 | | CRYSTAL STRUCTURE OF UNLIGANDED S. AUREUS THYMIDYLATE KINASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDYLATE KINASE | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
2CCJ
 
 | | Crystal structure of S. aureus thymidylate kinase complexed with thymidine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
1FKY
 
 | | NMR STUDY OF B-DNA CONTAINING A MISMATCHED BASE PAIR IN THE 29-39 K-RAS GENE SEQUENCE: CC CT C+C C+T, 2 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*CP*AP*GP*CP*TP*C)-3') | | Authors: | Boulard, Y, Cognet, J.A.H, Fazakerley, G.V. | | Deposit date: | 1996-10-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure as a function of pH of two central mismatches, C . T and C . C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics.
J.Mol.Biol., 268, 1997
|
|
1JHI
 
 | | Solution Structure of a Hedamycin-DNA complex | | Descriptor: | 5'-D(*AP*CP*CP*(HEH)GP*GP*T)-3', HEDAMYCIN | | Authors: | Owen, E.A, Burley, G.A, Carver, J.A, Wickham, G, Keniry, M.A. | | Deposit date: | 2001-06-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the hedamycin:d(ACCGGT)2 complex by NMR and restrained molecular dynamics.
Biochem.Biophys.Res.Commun., 290, 2002
|
|
1JUU
 
 | | NMR Structure of a Parallel Stranded DNA Duplex at Atomic Resolution | | Descriptor: | 5'-D(P*CP*CP*AP*TP*AP*AP*TP*TP*TP*AP*CP*C)-3', 5'-D(P*CP*CP*TP*AP*TP*TP*AP*AP*AP*TP*CP*C)-3' | | Authors: | Parvathy, V.R, Bhaumik, S.R, Chary, K.V.R, Govil, G, Liu, K, Howard, F.B, Miles, H.T. | | Deposit date: | 2001-08-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a parallel-stranded DNA duplex at atomic resolution.
Nucleic Acids Res., 30, 2002
|
|
1DNS
 
 | |
121D
 
 | | MOLECULAR STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR GROOVE BINDING DRUG NETROPSIN | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), NETROPSIN | | Authors: | Tabernero, L, Verdaguer, N, Coll, M, Fita, I, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Aymami, J. | | Deposit date: | 1993-04-14 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor groove binding drug netropsin.
Biochemistry, 32, 1993
|
|
7U8B
 
 | | Product of 14mer primer with activated asymmetric GA dimer diastereomer 1 | | Descriptor: | 5'-O-[(S)-(2-amino-1H-imidazol-1-yl)(hydroxy)phosphoryl]adenosine, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3'), SULFATE ION | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Understanding the metal ion function in the RNA non-enzymatic extesion with phosphorothioates
To Be Published
|
|
298D
 
 | | TARGETING THE MINOR GROOVE OF DNA: CRYSTAL STRUCTURES OF TWO COMPLEXES BETWEEN FURAN DERIVATIVES OF BERENIL AND THE DNA DODECAMER D(CGCGAATTCGCG)2 | | Descriptor: | 2,5-BIS{[4-(N-ISOPROPYL)DIAMINOMETHYL]PHENYL}FURAN, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Trent, J.O, Clark, G.R, Kumar, A, Wilson, W.D, Boykin, D.W, Hall, J.E, Tidwell, R.R, Blagburn, B.L, Neidle, S. | | Deposit date: | 1996-10-10 | | Release date: | 1996-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting the minor groove of DNA: crystal structures of two complexes between furan derivatives of berenil and the DNA dodecamer d(CGCGAATTCGCG)2.
J.Med.Chem., 39, 1996
|
|
289D
 
 | | TARGETING THE MINOR GROOVE OF DNA: CRYSTAL STRUCTURES OF TWO COMPLEXES BETWEEN FURAN DERIVATIVES OF BERENIL AND THE DNA DODECAMER D(CGCGAATTCGCG)2 | | Descriptor: | 2,5-BIS{[4-(N-CYCLOPROPYLDIAMINOMETHYL)PHENYL]}FURAN, DNA (5'-R(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Trent, J.O, Clark, G.R, Kumar, A, Wilson, W.D, Boykin, D.W, Hall, J.E, Tidwell, R.R, Blagburn, B.L, Neidle, S. | | Deposit date: | 1996-10-10 | | Release date: | 1996-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting the minor groove of DNA: crystal structures of two complexes between furan derivatives of berenil and the DNA dodecamer d(CGCGAATTCGCG)2.
J.Med.Chem., 39, 1996
|
|
3I2O
 
 | |
3I3M
 
 | |
3I49
 
 | |
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1O
 
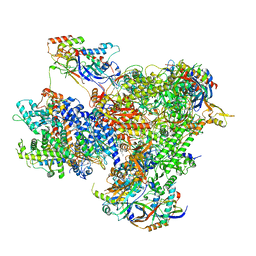 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
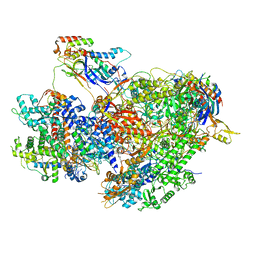 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1M
 
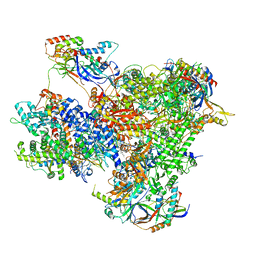 | | Structure of yeast RNA Polymerase III Elongation Complex (EC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
2D7D
 
 | |
1JS5
 
 | | Solution Structure of dAAUAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*UP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-16 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1JS7
 
 | | Solution Structure of dAAUAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*UP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1JRV
 
 | | SOLUTION STRUCTURE OF DAATAA DNA BULGE | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*TP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-15 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1JRW
 
 | | Solution Structure of dAATAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*TP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-15 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
4TRA
 
 | |
5SZX
 
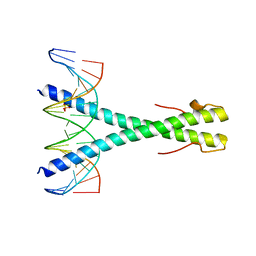 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5T01
 
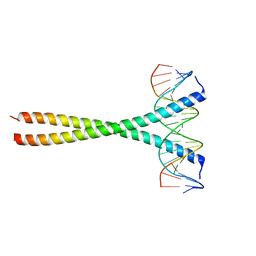 | | Human c-Jun DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*GP*AP*(5CM)P*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*GP*TP*CP*CP*AP*T)-3'), Transcription factor AP-1 | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
