2UZB
 
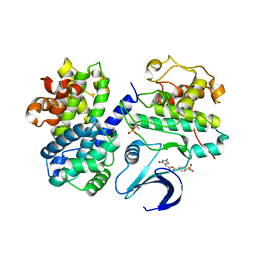 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 4-{5-[(Z)-(2-IMINO-4-OXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]-2-FURYL}-N-METHYLBENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5NU3
 
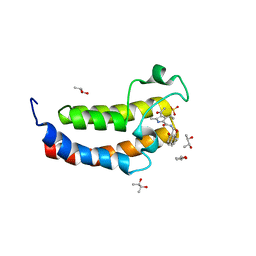 | | Crystal structure of the human bromodomain of CREBBP bound to the inhibitor XDM-CBP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CREB-binding protein, SULFATE ION, ... | | Authors: | Huegle, M, Wohlwend, D. | | Deposit date: | 2017-04-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8EPJ
 
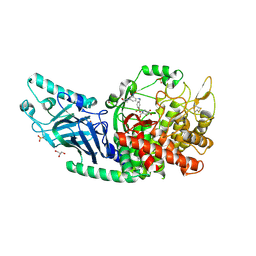 | | Co-crystal structure of Chaetomium glucosidase with compound 17 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[4-(morpholin-4-yl)-2-nitroanilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EHP
 
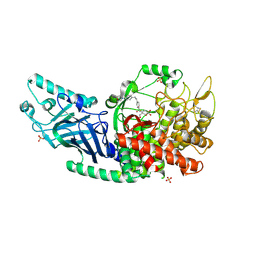 | | Co-crystal structure of Chaetomium glucosidase with compound 13 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(4-{[4-(morpholin-4-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8F17
 
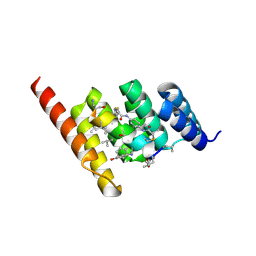 | | Structure of the STUB1 TPR domain in complex with H204, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8EPR
 
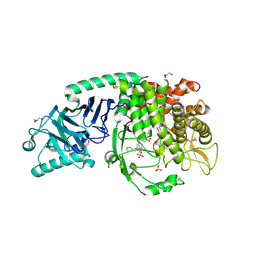 | | Co-crystal structure of Chaetomium glucosidase with compound 19 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[3-({[(5M)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EID
 
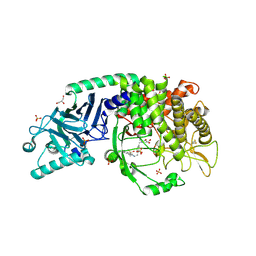 | | Co-crystal structure of Chaetomium glucosidase with compound 14 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({[(5P)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ELE
 
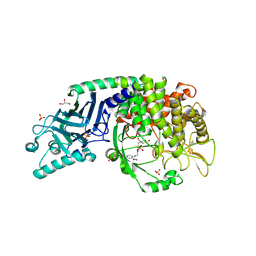 | | Co-crystal structure of Chaetomium glucosidase with compound 16 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-23 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
1KR3
 
 | | Crystal Structure of the Metallo beta-Lactamase from Bacteroides fragilis (CfiA) in Complex with the Tricyclic Inhibitor SB-236050. | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Payne, D.J, Hueso-Rodrguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Cheever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Dez, E, Prez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-beta-lactamases
ANTIMICROB.AGENTS CHEMOTHER., 46, 2002
|
|
1YF3
 
 | | T4Dam in Complex with AdoHcy and 13-mer Oligonucleotide Making Non- and Semi-specific (~1/4) Contact | | Descriptor: | 5'-D(*AP*CP*CP*AP*TP*GP*AP*TP*CP*TP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*CP*AP*TP*GP*G)-3', DNA adenine methylase, ... | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2004-12-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
8EUR
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 26 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EUT
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 27 | | Descriptor: | (2R,3R,4R,5S)-1-[8-(furan-2-yl)octyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
1KS8
 
 | | The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 2.5. | | Descriptor: | Endo-b-1,4-glucanase, SULFATE ION | | Authors: | Khademi, S, Guarino, L.A, Watanabe, H, Tokuda, G, Meyer, E.F. | | Deposit date: | 2002-01-11 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of an endoglucanase from termite, Nasutitermes takasagoensis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
2UZN
 
 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 4-{5-[(1Z)-1-(2-IMINO-4-OXO-1,3-THIAZOLIDIN-5-YLIDENE)ETHYL]-2-FURYL}BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4CH5
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated propionyl lysine | | Descriptor: | (S)-2-amino-6-propionamidohexanoic(((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
8EQ7
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 20 | | Descriptor: | (2R,3R,4R,5S)-1-[(3-{[3-bromo-5-(methanesulfonyl)anilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ER4
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 23 | | Descriptor: | (2R,3R,4R,5S)-1-(2-{6-[2-(4-azido-2-nitroanilino)ethyl]pyrazin-2-yl}ethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
3D7H
 
 | |
8F16
 
 | | Structure of the STUB1 TPR domain in complex with H203, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
5IJG
 
 | | Crystal structure of O-acetylhomoserine sulfhydrolase from Brucella melitensis at 2.0 A resolution | | Descriptor: | Cys/Met metabolism pyridoxal-phosphate-dependent enzyme, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Koolikova, V.V, Kotlov, M.I, Demidkina, T.V, Popov, V.O. | | Deposit date: | 2016-03-02 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of O-acetylhomoserine sulfhydrolase from Brucella melitensis at 2.0 A resolution
To Be Published
|
|
8F10
 
 | | Structure of the MDM2 P53 binding domain in complex with H102, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
9FCS
 
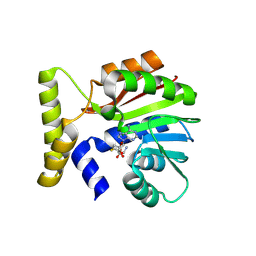 | | CysG(N-16)-H122N mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
9FCL
 
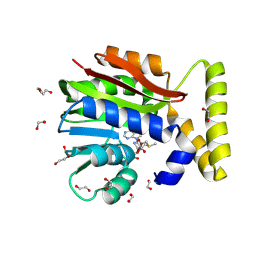 | | CysG(N-16) in complex with SAM from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
1Y7N
 
 | | Solution structure of the second PDZ domain of the human neuronal adaptor X11alpha | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 1 | | Authors: | Duquesne, A.E, de Ruijter, M, Brouwer, J, Drijfhout, J.W, Nabuurs, S.B, Spronk, C.A.E.M, Vuister, G.W, Ubbink, M, Canters, G.W. | | Deposit date: | 2004-12-09 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of the neuronal adaptor X11alpha and its interaction with the C-terminal peptide of the human copper chaperone for superoxide dismutase
J.Biomol.Nmr, 32, 2005
|
|
