5W1V
 
 | | Structure of the HLA-E-VMAPRTLIL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-04 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
7VPR
 
 | |
7VPU
 
 | |
5OJG
 
 | | Crystal structure of the dehydrogenase/reductase SDR family member 4 (DHRS4) from Caenorhabditis elegans | | Descriptor: | Dehydrogenase/reductase SDR family member 4, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, butane-2,3-dione | | Authors: | Scheidig, A.J, Faust, A, Ebert, B, Maser, E, Kisiela, M. | | Deposit date: | 2017-07-21 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and catalytic characterization of the dehydrogenase/reductase SDR family member 4 (DHRS4) from Caenorhabditis elegans.
FEBS J., 285, 2018
|
|
5O0Y
 
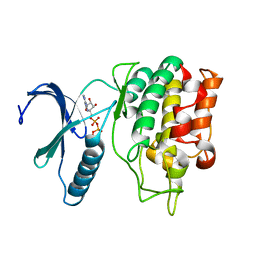 | | TLK2 kinase domain from human | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase tousled-like 2 | | Authors: | Mortuza, G.B, Montoya, G. | | Deposit date: | 2017-05-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis of Tousled-Like Kinase 2 activation.
Nat Commun, 9, 2018
|
|
7V4Q
 
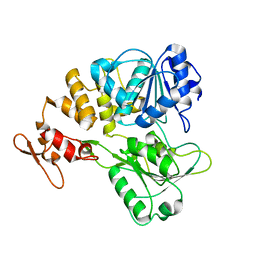 | |
5OLY
 
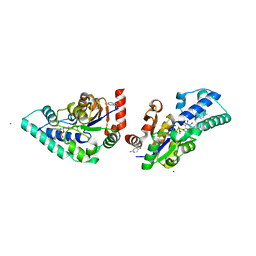 | |
5VL4
 
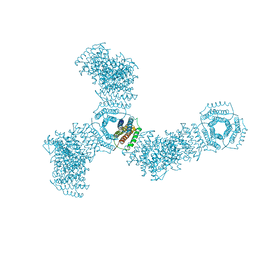 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
5VJU
 
 | | De Novo Photosynthetic Reaction Center Protein Variant Equipped with His-Tyr H-bond, Heme B, and Cd(II) ions | | Descriptor: | CADMIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette Leu71His variant | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
8BBI
 
 | |
6J62
 
 | | Crystal structure of mISG15/NS1B complex | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Jiang, Y.N, Wang, X.Q. | | Deposit date: | 2019-01-12 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Crystal structure of mISG15/NS1B complex
To Be Published
|
|
5O51
 
 | | AfRom2 CNH domain | | Descriptor: | Rho guanyl nucleotide exchange factor (Rom2), putative | | Authors: | Wei, W, van Aalten, D. | | Deposit date: | 2017-05-31 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rom2 CNH domain from Aspergillus fumigatus is an atypical seven-bladed WD-40 protein
To Be Published
|
|
5KGX
 
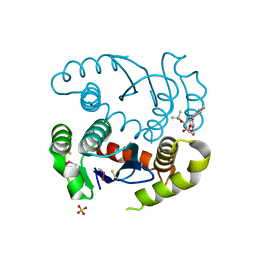 | | HIV1 catalytic core domain in complex with an inhibitor (2~{S})-2-[3-(3,4-dihydro-2~{H}-chromen-6-yl)-1-methyl-indol-2-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid | | Descriptor: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-methyl-1H-indol-2-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Feng, L, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-10-19 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Indole-based allosteric inhibitors of HIV-1 integrase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6RMM
 
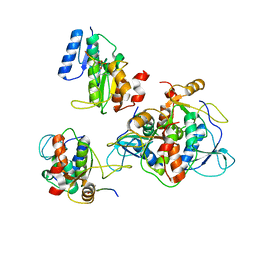 | |
6XO9
 
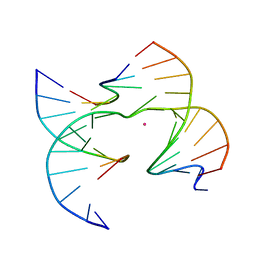 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J33 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*AP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6J91
 
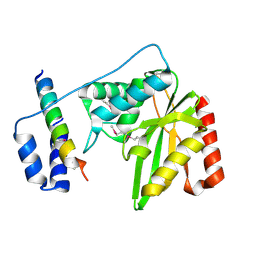 | | Structure of a hypothetical protease | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VPG
 
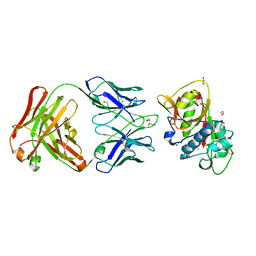 | | CRYSTAL STRUCTURE OF DER P 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
6XZS
 
 | | Crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((4-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(4-fluorophenyl)methylamino]ethyl]-3-(4-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, F, Supuran, C.T. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
6XZY
 
 | | crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((2-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(2-fluorophenyl)methylamino]ethyl]-3-(3-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
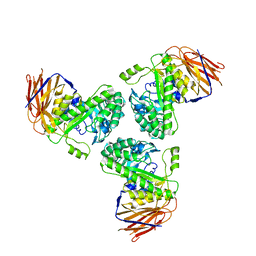
5O82
 
 | |
8BG2
 
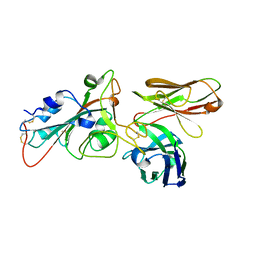 | |
5O90
 
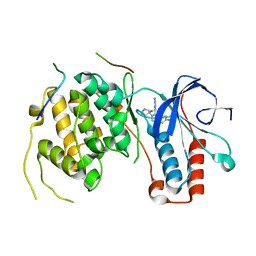 | | Crystal structure of a P38alpha T185G mutant in complex with TAB1 peptide. | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, Mitogen-activated protein kinase 14, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Nichols, C.E, De Nicola, G.F, Thapa, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | TAB1-Induced Autoactivation of p38 alpha Mitogen-Activated Protein Kinase Is Crucially Dependent on Threonine 185.
Mol. Cell. Biol., 38, 2018
|
|
7VKA
 
 | | Crystal Structure of GH3.6 in complex with an inhibitor | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | Authors: | Wang, N, Luo, M, Bao, H, Huang, H. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KL6
 
 | | Wilms Tumor Protein (WT1) Q369R ZnF2-4 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
