8THD
 
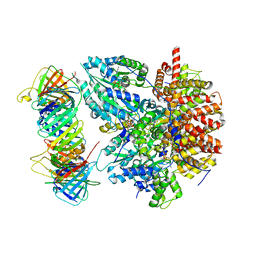 | | Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to PCNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | Authors: | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-07-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
8THC
 
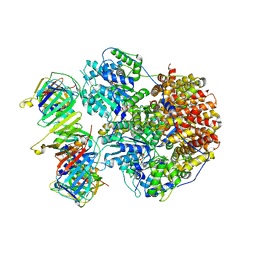 | | Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to a cracked PCNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | Authors: | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-07-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
7FIL
 
 | |
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | Descriptor: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | Authors: | Shi, Y, Huang, G, Zhan, X. | | Deposit date: | 2021-07-31 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
3QQC
 
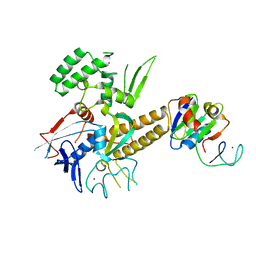 | | Crystal structure of archaeal Spt4/5 bound to the RNAP clamp domain | | Descriptor: | DNA-directed RNA polymerase subunit b, DNA-directed RNA polymerase subunit a', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Martinez-Rucobo, F.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity.
Embo J., 30, 2011
|
|
6S2F
 
 | |
6S2E
 
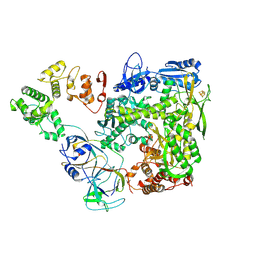 | |
6S1C
 
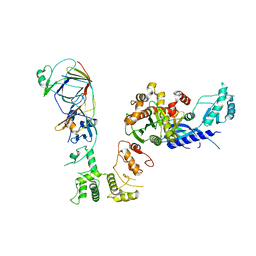 | | P3221 crystal form of the Ctf18-1-8/Pol2(1-528) complex | | Descriptor: | Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Grabarczyk, D.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (6.1 Å) | | Cite: | Ctf18-RFC and DNA Pol ε form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication.
Nucleic Acids Res., 48, 2020
|
|
4X1B
 
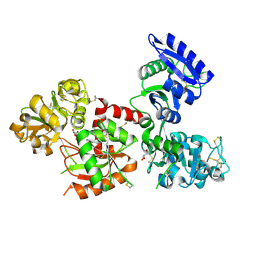 | | Human serum transferrin with ferric ion bound at the C-lobe only | | Descriptor: | FE (III) ION, GLYCEROL, MALONATE ION, ... | | Authors: | Wang, M, Zhang, H, Sun, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | "Anion clamp" allows flexible protein to impose coordination geometry on metal ions
Chem.Commun.(Camb.), 51, 2015
|
|
2JPP
 
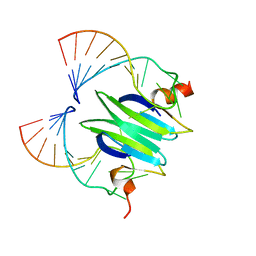 | | Structural basis of RsmA/CsrA RNA recognition: Structure of RsmE bound to the Shine-Dalgarno sequence of hcnA mRNA | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*UP*CP*AP*CP*GP*GP*AP*UP*GP*AP*AP*GP*CP*CP*C)-3'), Translational repressor | | Authors: | Schubert, M, Lapouge, K, Duss, O, Oberstrass, F.C, Jelesarov, I, Haas, D, Allain, F.H.-T. | | Deposit date: | 2007-05-21 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA
Nat.Struct.Mol.Biol., 14, 2007
|
|
1RXZ
 
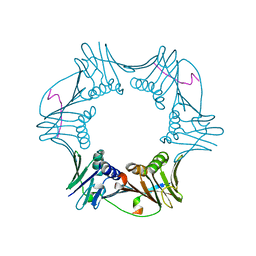 | | C-terminal region of A. fulgidus FEN-1 complexed with A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
6KNB
 
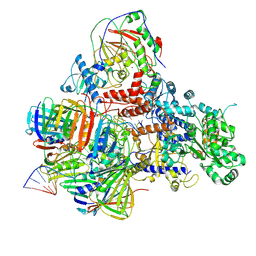 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
3TXN
 
 | |
3TXM
 
 | |
6KNC
 
 | | PolD-PCNA-DNA (form B) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
7AIB
 
 | | MutS-MutL in clamp state | | Descriptor: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8TW8
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA complex in Apo state conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
8TW7
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA complex in Apo state conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
8TWB
 
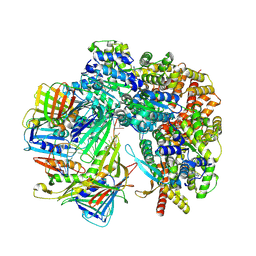 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
8TW9
 
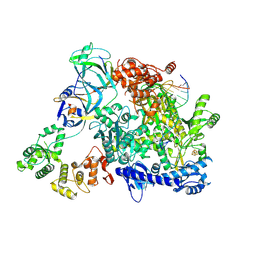 | | Cryo-EM structure of S. cerevisiae PolE-Ctf18-8-1-DNA | | Descriptor: | Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
8TWA
 
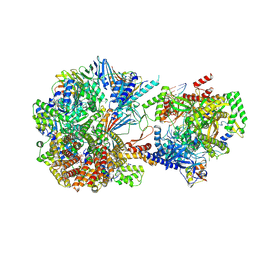 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-PolE-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
7AUU
 
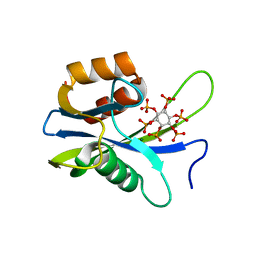 | |
7AUT
 
 | |
7AUN
 
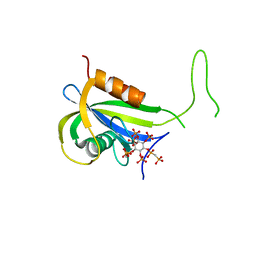 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PCP-InsP8 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, MAGNESIUM ION, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(hydroxyphosphoryl)methanediyl]}bis(phosphonic acid) | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUO
 
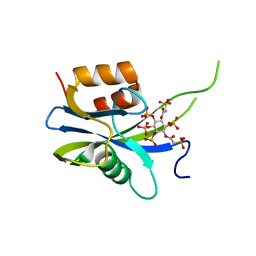 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PA-InsP8 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(2-oxoethane-2,1-diyl)]}bis(phosphonic acid) | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
