4I4B
 
 | | HMG-CoA Reductase from Pseudomonas mevalonii complexed with NAD and intermediate hemiacetal form of HMG-CoA | | Descriptor: | (3R,5R,9R,19R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8,21-trimethyl-10,14-dioxo-19-sulfanyl-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, (3S)-3-hydroxy-3-methyl-5-sulfanylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, ... | | Authors: | Stauffacher, C.V, Steussy, C.N, Critchelow, C.J, Schmidt, T, Min, J, Wrensford, L.V, Burgner, J.W, Rodwell, V.W. | | Deposit date: | 2012-11-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
1JQ5
 
 | | Bacillus Stearothermophilus Glycerol dehydrogenase complex with NAD+ | | Descriptor: | Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
3TNZ
 
 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) C217A, C239A bound to FMN and mono-iodotyrosine (MIT) | | Descriptor: | 3-IODO-TYROSINE, CITRATE ANION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Buss, J.M, McTamney, P.M, Rokita, S.E. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Expression of a soluble form of iodotyrosine deiodinase for active site characterization by engineering the native membrane protein from Mus musculus.
Protein Sci., 21, 2012
|
|
5RSN
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000064576 | | Descriptor: | (1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1RX4
 
 | |
3OCH
 
 | | Chemically Self-assembled Antibody Nanorings (CSANs): Design and Characterization of an Anti-CD3 IgM Biomimetic | | Descriptor: | (2S,2'S)-5,5'-(nonane-1,9-diyldiimino)bis(2-{[(4-{[(2,4-diaminopteridin-6-yl)methyl](methyl)amino}phenyl)carbonyl]amino}-5-oxopentanoic acid) (non-preferred name), CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cody, V. | | Deposit date: | 2010-08-10 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Chemically Self-Assembled Antibody Nanorings (CSANs): Design and Characterization of an Anti-CD3 IgM Biomimetic.
J.Am.Chem.Soc., 132, 2010
|
|
1WZI
 
 | |
3G4Z
 
 | | Crystal Structure of NiSOD Y9F mutant at 1.9 A | | Descriptor: | BROMIDE ION, NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Garman, S.C, Guce, A.I, Herbst, R.W, Bryngelson, P.A, Cabelli, D.E, Higgins, K.A, Ryan, K.C, Maroney, M.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution
Biochemistry, 48, 2009
|
|
1IXB
 
 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE(II) SUPEROXIDE DISMUTASE MUTANT Y174F AT 0.90 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE ION, 1 HYDROXYL COORDINATED, SUPEROXIDE DISMUTASE | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-18 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
3ZCH
 
 | | Ascorbate peroxidase W41A-H42M mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASCORBATE PEROXIDASE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Conformational Mobility of the Active Site of Ascorbate Peroxidase
Dalton Trans, 42, 2013
|
|
3L68
 
 | | Xenobiotic Reductase A - C25S variant with coumarin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
3G1R
 
 | | Crystal structure of human liver 5beta-reductase (AKR1D1) in complex with NADP and Finasteride. Resolution 1.70 A | | Descriptor: | (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide, 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J.E, Penning, T.M, Christianson, D.W. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Inhibition of human steroid 5beta-reductase (AKR1D1) by finasteride and structure of the enzyme-inhibitor complex.
J.Biol.Chem., 284, 2009
|
|
1RX6
 
 | |
3BLV
 
 | | Yeast Isocitrate Dehydrogenase with Citrate Bound in the Regulatory Subunits | | Descriptor: | CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, Isocitrate dehydrogenase [NAD] subunit 2 | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
1RX5
 
 | |
1RX9
 
 | |
3SQP
 
 | | Structure of human glutathione reductase complexed with pyocyanin, an agent with antimalarial activity | | Descriptor: | 5-methylphenazin-1(5H)-one, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fritz-Wolf, K, Schirmer, R.H, Koenig, I, Goebel, U. | | Deposit date: | 2011-07-06 | | Release date: | 2011-09-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The bacterial redox signaller pyocyanin as an antiplasmodial agent: comparisons with its thioanalog methylene blue.
Redox Rep., 16, 2011
|
|
5AOG
 
 | | Structure of Sorghum peroxidase | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, CALCIUM ION, CATIONIC PEROXIDASE SPC4, ... | | Authors: | Kwon, H, Nnamchi, C.I, Parkin, G, Efimov, I, Agirre, J, Basran, J, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2015-09-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural and Spectroscopic Characterisation of a Heme Peroxidase from Sorghum.
J.Biol.Inorg.Chem., 21, 2016
|
|
1AUJ
 
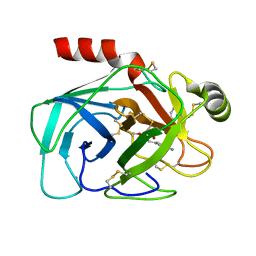 | | BOVINE TRYPSIN COMPLEXED TO META-CYANO-BENZYLIC INHIBITOR | | Descriptor: | 1-{[1-(2-AMINO-3-PHENYL-PROPIONYL)-PYRROLIDINE-2-CARBONYL]-AMINO}-2-(3-CYANO-PHENYL)-ETHANEBORONIC ACID, CALCIUM ION, TRYPSIN | | Authors: | Alexander, R, Smallwood, A, Kettner, C. | | Deposit date: | 1997-08-28 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New inhibitors of thrombin and other trypsin-like proteases: hydrogen bonding of an aromatic cyano group with a backbone amide of the P1 binding site replaces binding of a basic side chain.
Biochemistry, 36, 1997
|
|
3CN3
 
 | |
3CUR
 
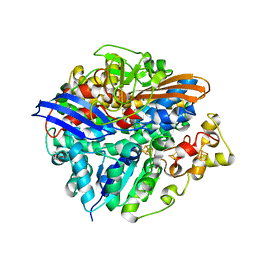 | | Structure of a double methionine mutant of NI-FE hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A. | | Deposit date: | 2008-04-17 | | Release date: | 2008-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental approaches to kinetics of gas diffusion in hydrogenase
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3P7X
 
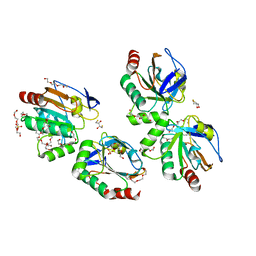 | | Crystal structure of an atypical two-cysteine peroxiredoxin (SAOUHSC_01822) from Staphylococcus aureus NCTC8325 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Probable thiol peroxidase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2010-10-13 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of an atypical two-cysteine peroxiredoxin (SAOUHSC_01822) from Staphylococcus aureus NCTC8325
To be Published
|
|
5ZCQ
 
 | | Azide-bound cytochrome c oxidase structure determined using the crystals exposed to 10 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
2D3Q
 
 | |
5Z84
 
 | | The structure of azide-bound cytochrome c oxidase determined using the crystals exposed to 20 mM azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
